Note
Go to the end to download the full example code.
Petrophysically guided inversion (PGI): Linear example#
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically constrained inversion. We explore it through the UBC linear example.
Tikhonov Inversion##
import discretize as Mesh
import matplotlib.pyplot as plt
import numpy as np
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 20
jk = np.linspace(1.0, 60.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
# True model
mtrue = np.zeros(mesh.nC)
mtrue[mesh.cell_centers_x > 0.2] = 1.0
mtrue[mesh.cell_centers_x > 0.35] = 0.0
t = (mesh.cell_centers_x - 0.65) / 0.25
indx = np.abs(t) < 1
mtrue[indx] = -(((1 - t**2.0) ** 2.0)[indx])
mtrue = np.zeros(mesh.nC)
mtrue[mesh.cell_centers_x > 0.3] = 1.0
mtrue[mesh.cell_centers_x > 0.45] = -0.5
mtrue[mesh.cell_centers_x > 0.6] = 0
# simpeg problem and survey
prob = simulation.LinearSimulation(mesh, G=G, model_map=maps.IdentityMap())
std = 0.01
survey = prob.make_synthetic_data(mtrue, relative_error=std, add_noise=True)
# Setup the inverse problem
reg = regularization.WeightedLeastSquares(mesh, alpha_s=1.0, alpha_x=1.0)
dmis = data_misfit.L2DataMisfit(data=survey, simulation=prob)
opt = optimization.ProjectedGNCG(maxIter=10, cg_maxiter=50, cg_rtol=1e-3)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
directiveslist = [
directives.BetaEstimate_ByEig(beta0_ratio=1e-5),
directives.BetaSchedule(coolingFactor=10.0, coolingRate=2),
directives.TargetMisfit(),
]
inv = inversion.BaseInversion(invProb, directiveList=directiveslist)
m0 = np.zeros_like(mtrue)
mnormal = inv.run(m0)
Running inversion with SimPEG v0.25.2.dev2+gfcb9bdf36
================================================= Projected GNCG =================================================
# beta phi_d phi_m f |proj(x-g)-x| LS iter_CG CG |Ax-b|/|b| CG |Ax-b| Comment
-----------------------------------------------------------------------------------------------------------------
0 1.81e+01 2.00e+05 0.00e+00 2.00e+05 0 inf inf
1 1.81e+01 3.18e+02 4.26e+01 1.09e+03 2.49e+06 0 13 7.18e-04 1.79e+03
2 1.81e+01 3.58e+01 4.59e+01 8.67e+02 1.79e+03 0 21 4.34e-04 7.77e-01 Skip BFGS
3 1.81e+00 5.64e+00 4.92e+01 9.48e+01 5.03e+02 0 32 1.68e-04 8.47e-02 Skip BFGS
------------------------- STOP! -------------------------
1 : |fc-fOld| = 2.4109e+01 <= tolF*(1+|f0|) = 2.0000e+04
0 : |xc-x_last| = 1.8797e-01 <= tolX*(1+|x0|) = 1.0000e-01
0 : |proj(x-g)-x| = 5.0344e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 5.0344e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 10 <= iter = 3
------------------------- DONE! -------------------------
Petrophysically constrained inversion ##
# fit a Gaussian Mixture Model with n components
# on the true model to simulate the laboratory
# petrophysical measurements
n = 3
clf = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=n,
covariance_type="full",
max_iter=100,
n_init=3,
reg_covar=5e-4,
)
clf.fit(mtrue.reshape(-1, 1))
# Petrophyically constrained regularization
reg = regularization.PGI(
gmmref=clf,
mesh=mesh,
alpha_pgi=1.0,
alpha_x=1.0,
)
# Optimization
opt = optimization.ProjectedGNCG(maxIter=20, cg_maxiter=50, cg_rtol=1e-3)
opt.remember("xc")
# Setup new inverse problem
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
Alphas = directives.AlphasSmoothEstimate_ByEig(alpha0_ratio=10.0, verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-8)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
warmingFactor=1.0,
tolerance=0.1,
update_rate=1,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters()
addmref = directives.PGI_AddMrefInSmooth(verbose=True)
# Setup Inversion
inv = inversion.BaseInversion(
invProb, directiveList=[Alphas, beta, petrodir, targets, addmref, betaIt]
)
# Initial model same as for WeightedLeastSquares
mcluster = inv.run(m0)
# Final Plot
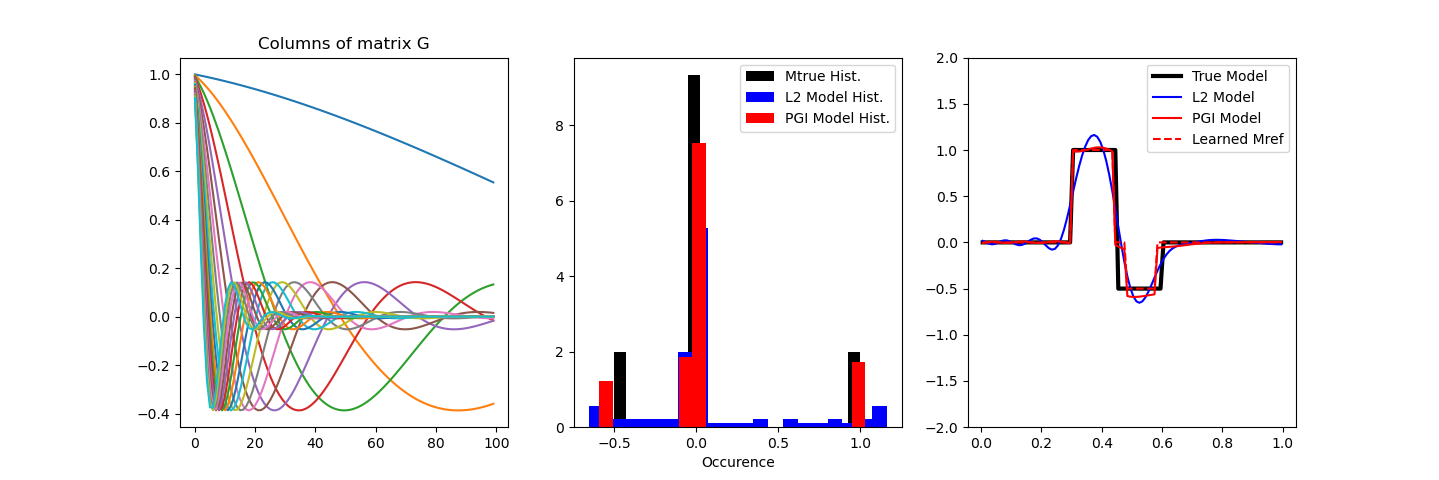
fig, axes = plt.subplots(1, 3, figsize=(12 * 1.2, 4 * 1.2))
for i in range(prob.G.shape[0]):
axes[0].plot(prob.G[i, :])
axes[0].set_title("Columns of matrix G")
axes[1].hist(mtrue, bins=20, linewidth=3.0, density=True, color="k")
axes[1].set_xlabel("Model value")
axes[1].set_xlabel("Occurence")
axes[1].hist(mnormal, bins=20, density=True, color="b")
axes[1].hist(mcluster, bins=20, density=True, color="r")
axes[1].legend(["Mtrue Hist.", "L2 Model Hist.", "PGI Model Hist."])
axes[2].plot(mesh.cell_centers_x, mtrue, color="black", linewidth=3)
axes[2].plot(mesh.cell_centers_x, mnormal, color="blue")
axes[2].plot(mesh.cell_centers_x, mcluster, "r-")
axes[2].plot(mesh.cell_centers_x, invProb.reg.objfcts[0].reference_model, "r--")
axes[2].legend(("True Model", "L2 Model", "PGI Model", "Learned Mref"))
axes[2].set_ylim([-2, 2])
plt.show()
Running inversion with SimPEG v0.25.2.dev2+gfcb9bdf36
Alpha scales: [np.float64(0.5236110258717401), np.float64(0.0)]
<class 'simpeg.regularization.pgi.PGIsmallness'>
================================================= Projected GNCG =================================================
# beta phi_d phi_m f |proj(x-g)-x| LS iter_CG CG |Ax-b|/|b| CG |Ax-b| Comment
-----------------------------------------------------------------------------------------------------------------
0 3.16e-02 2.00e+05 0.00e+00 2.00e+05 0 inf inf
1 3.16e-02 4.06e+01 3.54e+02 5.18e+01 2.49e+06 0 14 5.06e-04 1.26e+03
geophys. misfits: 40.6 (target 20.0 [False]) | smallness misfit: 2978.3 (target: 100.0 [False])
mref changed in 26 places
Beta cooling evaluation: progress: [40.6]; minimum progress targets: [160000.]
2 3.16e-02 4.94e+00 5.22e+01 6.59e+00 1.26e+03 0 36 2.71e-04 3.42e-01 Skip BFGS
geophys. misfits: 4.9 (target 20.0 [True]) | smallness misfit: 2039.3 (target: 100.0 [False])
mref changed in 1 places
Beta cooling evaluation: progress: [4.9]; minimum progress targets: [32.5]
Warming alpha_pgi to favor clustering: 4.050532807512921
3 3.16e-02 5.02e+00 8.38e+01 7.67e+00 4.26e+00 0 50 9.06e-01 3.86e+00
geophys. misfits: 5.0 (target 20.0 [True]) | smallness misfit: 843.2 (target: 100.0 [False])
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
Beta cooling evaluation: progress: [5.]; minimum progress targets: [22.]
Warming alpha_pgi to favor clustering: 16.140848516325573
4 3.16e-02 5.17e+00 1.39e+02 9.55e+00 1.07e+01 0 50 1.57e+00 1.68e+01
geophys. misfits: 5.2 (target 20.0 [True]) | smallness misfit: 333.1 (target: 100.0 [False])
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
Beta cooling evaluation: progress: [5.2]; minimum progress targets: [22.]
Warming alpha_pgi to favor clustering: 62.47977485677254
5 3.16e-02 5.79e+00 2.18e+02 1.27e+01 3.03e+01 0 50 1.52e+00 4.60e+01
geophys. misfits: 5.8 (target 20.0 [True]) | smallness misfit: 164.2 (target: 100.0 [False])
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
Beta cooling evaluation: progress: [5.8]; minimum progress targets: [22.]
Warming alpha_pgi to favor clustering: 215.9269598795071
6 3.16e-02 7.71e+00 3.33e+02 1.82e+01 7.87e+01 0 50 1.32e+00 1.04e+02
geophys. misfits: 7.7 (target 20.0 [True]) | smallness misfit: 94.5 (target: 100.0 [True])
All targets have been reached
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
Beta cooling evaluation: progress: [7.7]; minimum progress targets: [22.]
Warming alpha_pgi to favor clustering: 560.1644488563037
------------------------- STOP! -------------------------
1 : |fc-fOld| = 2.4288e+00 <= tolF*(1+|f0|) = 2.0000e+04
0 : |xc-x_last| = 1.0947e-01 <= tolX*(1+|x0|) = 1.0000e-01
0 : |proj(x-g)-x| = 7.8744e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 7.8744e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 20 <= iter = 6
------------------------- DONE! -------------------------
Total running time of the script: (0 minutes 4.262 seconds)
Estimated memory usage: 321 MB
