Note
Go to the end to download the full example code.
Joint PGI of Gravity + Magnetic on an Octree mesh using full petrophysical information#
This tutorial shows through a joint inversion of Gravity and Magnetic data on an Octree mesh how to use the PGI framework introduced in Astic & Oldenburg (2019) and Astic et al. (2021) to include petrophysical information into geophysical inversions for mutli-physics inversion.
Thibaut Astic, Douglas W. Oldenburg, A framework for petrophysically and geologically guided geophysical inversion using a dynamic Gaussian mixture model prior, Geophysical Journal International, Volume 219, Issue 3, December 2019, Pages 1989–2012, DOI: 10.1093/gji/ggz389.
Thibaut Astic, Lindsey J. Heagy, Douglas W Oldenburg, Petrophysically and geologically guided multi-physics inversion using a dynamic Gaussian mixture model, Geophysical Journal International, Volume 224, Issue 1, January 2021, Pages 40-68, DOI: 10.1093/gji/ggaa378.
Import modules#
from discretize import TreeMesh
from discretize.utils import active_from_xyz
import matplotlib.pyplot as plt
import numpy as np
import simpeg.potential_fields as pf
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
utils,
)
from simpeg.utils import io_utils
# Reproducible science
np.random.seed(518936)
Setup#
# Load Mesh
mesh_file = io_utils.download(
"https://storage.googleapis.com/simpeg/pgi_tutorial_assets/mesh_tutorial.ubc"
)
mesh = TreeMesh.read_UBC(mesh_file)
# Load True geological model for comparison with inversion result
true_geology_file = io_utils.download(
"https://storage.googleapis.com/simpeg/pgi_tutorial_assets/geology_true.mod"
)
true_geology = mesh.read_model_UBC(true_geology_file)
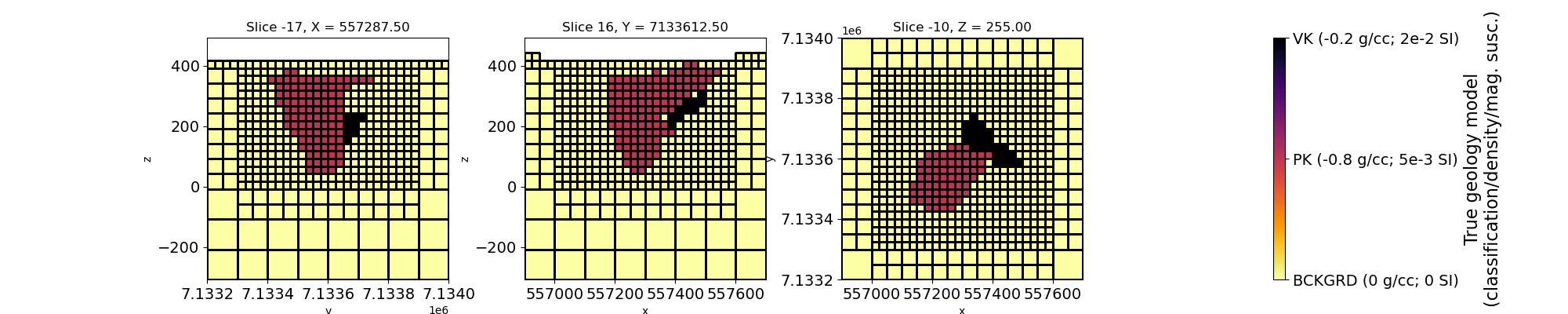
# Plot true geology model
fig, ax = plt.subplots(1, 4, figsize=(20, 4))
ticksize, labelsize = 14, 16
for _, axx in enumerate(ax):
axx.set_aspect(1)
axx.tick_params(labelsize=ticksize)
mesh.plot_slice(
true_geology,
normal="X",
ax=ax[0],
ind=-17,
clim=[0, 2],
pcolor_opts={"cmap": "inferno_r"},
grid=True,
)
mesh.plot_slice(
true_geology,
normal="Y",
ax=ax[1],
clim=[0, 2],
pcolor_opts={"cmap": "inferno_r"},
grid=True,
)
geoplot = mesh.plot_slice(
true_geology,
normal="Z",
ax=ax[2],
clim=[0, 2],
ind=-10,
pcolor_opts={"cmap": "inferno_r"},
grid=True,
)
geocb = plt.colorbar(geoplot[0], cax=ax[3], ticks=[0, 1, 2])
geocb.set_label(
"True geology model\n(classification/density/mag. susc.)", fontsize=labelsize
)
geocb.set_ticklabels(
["BCKGRD (0 g/cc; 0 SI)", "PK (-0.8 g/cc; 5e-3 SI)", "VK (-0.2 g/cc; 2e-2 SI)"]
)
geocb.ax.tick_params(labelsize=ticksize)
ax[3].set_aspect(10)
plt.show()
# Load geophysical data
data_grav_file = io_utils.download(
"https://storage.googleapis.com/simpeg/pgi_tutorial_assets/gravity_data.obs"
)
data_grav = io_utils.read_grav3d_ubc(data_grav_file)
data_mag_file = io_utils.download(
"https://storage.googleapis.com/simpeg/pgi_tutorial_assets/magnetic_data.obs"
)
data_mag = io_utils.read_mag3d_ubc(data_mag_file)
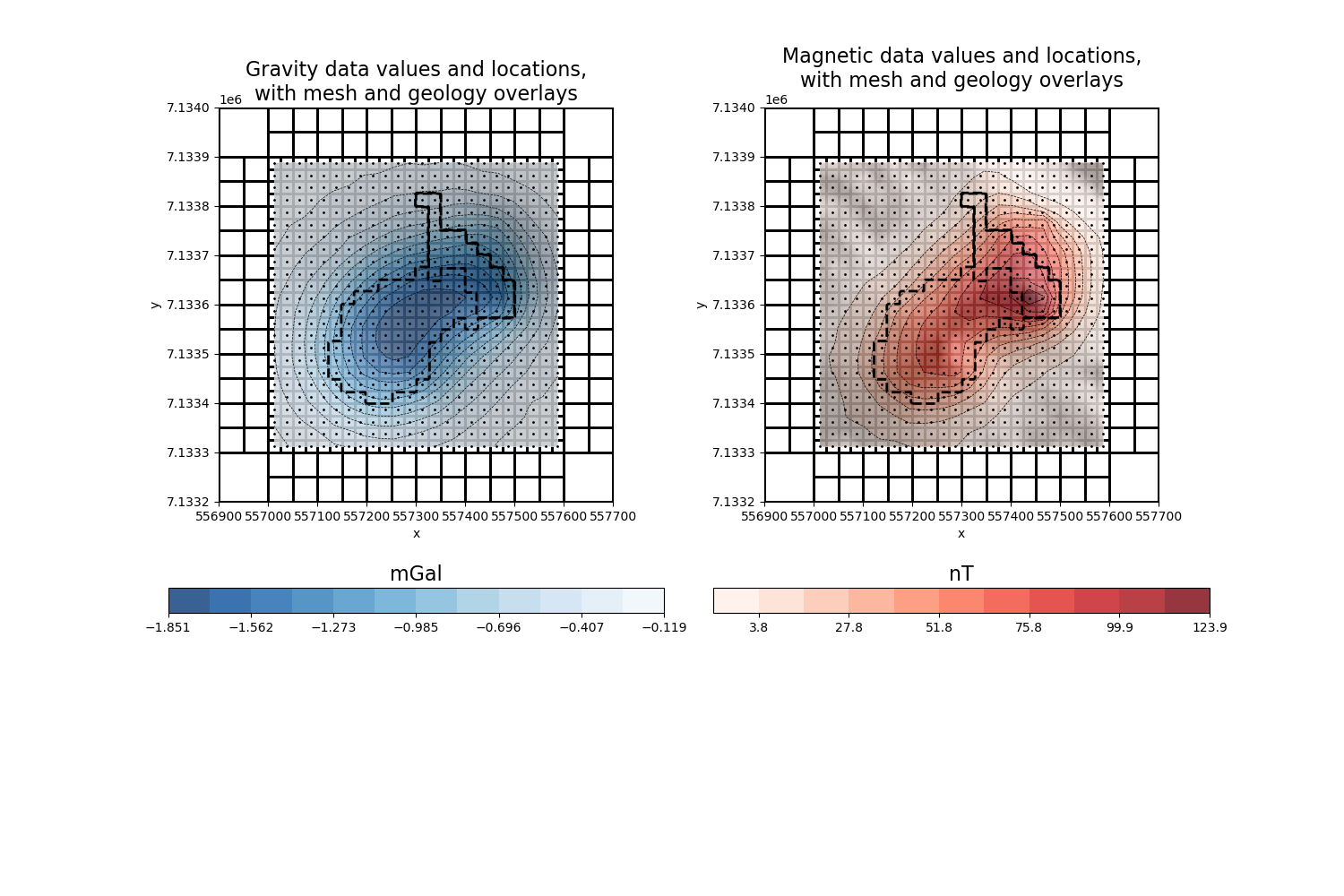
# plot data and mesh
fig, ax = plt.subplots(2, 2, figsize=(15, 10))
ax = ax.reshape(-1)
plt.gca().set_aspect("equal")
plt.gca().set_xlim(
[
data_mag.survey.receiver_locations[:, 0].min(),
data_mag.survey.receiver_locations[:, 0].max(),
],
)
plt.gca().set_ylim(
[
data_mag.survey.receiver_locations[:, 1].min(),
data_mag.survey.receiver_locations[:, 1].max(),
],
)
mesh.plot_slice(
np.ones(mesh.nC),
normal="Z",
ind=int(-10),
grid=True,
pcolor_opts={"cmap": "Greys"},
ax=ax[0],
)
mm = utils.plot2Ddata(
data_grav.survey.receiver_locations,
-data_grav.dobs,
ax=ax[0],
level=True,
nx=20,
ny=20,
dataloc=True,
ncontour=12,
shade=True,
contourOpts={"cmap": "Blues_r", "alpha": 0.8},
levelOpts={"colors": "k", "linewidths": 0.5, "linestyles": "dashed"},
)
ax[0].set_aspect(1)
ax[0].set_title(
"Gravity data values and locations,\nwith mesh and geology overlays", fontsize=16
)
plt.colorbar(mm[0], cax=ax[2], orientation="horizontal")
ax[2].set_aspect(0.05)
ax[2].set_title("mGal", fontsize=16)
mesh.plot_slice(
np.ones(mesh.nC),
normal="Z",
ind=int(-10),
grid=True,
pcolor_opts={"cmap": "Greys"},
ax=ax[1],
)
mm = utils.plot2Ddata(
data_mag.survey.receiver_locations,
data_mag.dobs,
ax=ax[1],
level=True,
nx=20,
ny=20,
dataloc=True,
ncontour=11,
shade=True,
contourOpts={"cmap": "Reds", "alpha": 0.8},
levelOpts={"colors": "k", "linewidths": 0.5, "linestyles": "dashed"},
)
ax[1].set_aspect(1)
ax[1].set_title(
"Magnetic data values and locations,\nwith mesh and geology overlays", fontsize=16
)
plt.colorbar(mm[0], cax=ax[3], orientation="horizontal")
ax[3].set_aspect(0.05)
ax[3].set_title("nT", fontsize=16)
# overlay true geology model for comparison
indz = -9
indslicezplot = mesh.gridCC[:, 2] == mesh.cell_centers_z[indz]
for i in range(2):
utils.plot2Ddata(
mesh.gridCC[indslicezplot][:, [0, 1]],
true_geology[indslicezplot],
nx=200,
ny=200,
contourOpts={"alpha": 0},
clim=[0, 2],
ax=ax[i],
level=True,
ncontour=2,
levelOpts={"colors": "k", "linewidths": 2, "linestyles": "--"},
method="nearest",
)
plt.subplots_adjust(hspace=-0.25, wspace=0.1)
plt.show()
# Load Topo
topo_file = io_utils.download(
"https://storage.googleapis.com/simpeg/pgi_tutorial_assets/CDED_Lake_warp.xyz"
)
topo = np.genfromtxt(topo_file, skip_header=1)
# find the active cells
actv = active_from_xyz(mesh, topo, "CC")
# Create active map to go from reduce set to full
ndv = np.nan
actvMap = maps.InjectActiveCells(mesh, actv, ndv)
nactv = int(actv.sum())
# Create simulations and data misfits
# Wires mapping
wires = maps.Wires(("den", actvMap.nP), ("sus", actvMap.nP))
gravmap = actvMap * wires.den
magmap = actvMap * wires.sus
idenMap = maps.IdentityMap(nP=nactv)
# Grav problem
simulation_grav = pf.gravity.simulation.Simulation3DIntegral(
survey=data_grav.survey,
mesh=mesh,
rhoMap=wires.den,
ind_active=actv,
engine="choclo",
)
dmis_grav = data_misfit.L2DataMisfit(data=data_grav, simulation=simulation_grav)
# Mag problem
simulation_mag = pf.magnetics.simulation.Simulation3DIntegral(
survey=data_mag.survey,
mesh=mesh,
chiMap=wires.sus,
ind_active=actv,
)
dmis_mag = data_misfit.L2DataMisfit(data=data_mag, simulation=simulation_mag)
Downloading https://storage.googleapis.com/simpeg/pgi_tutorial_assets/mesh_tutorial.ubc
saved to: /home/vsts/work/1/s/tutorials/14-pgi/mesh_tutorial.ubc
Download completed!
Downloading https://storage.googleapis.com/simpeg/pgi_tutorial_assets/geology_true.mod
saved to: /home/vsts/work/1/s/tutorials/14-pgi/geology_true.mod
Download completed!
Downloading https://storage.googleapis.com/simpeg/pgi_tutorial_assets/gravity_data.obs
saved to: /home/vsts/work/1/s/tutorials/14-pgi/gravity_data.obs
Download completed!
Downloading https://storage.googleapis.com/simpeg/pgi_tutorial_assets/magnetic_data.obs
saved to: /home/vsts/work/1/s/tutorials/14-pgi/magnetic_data.obs
Download completed!
Downloading https://storage.googleapis.com/simpeg/pgi_tutorial_assets/CDED_Lake_warp.xyz
saved to: /home/vsts/work/1/s/tutorials/14-pgi/CDED_Lake_warp.xyz
Download completed!
Create a joint Data Misfit#
Inversion with full petrophysical information#
Create and plot a petrophysical GMM with full information#
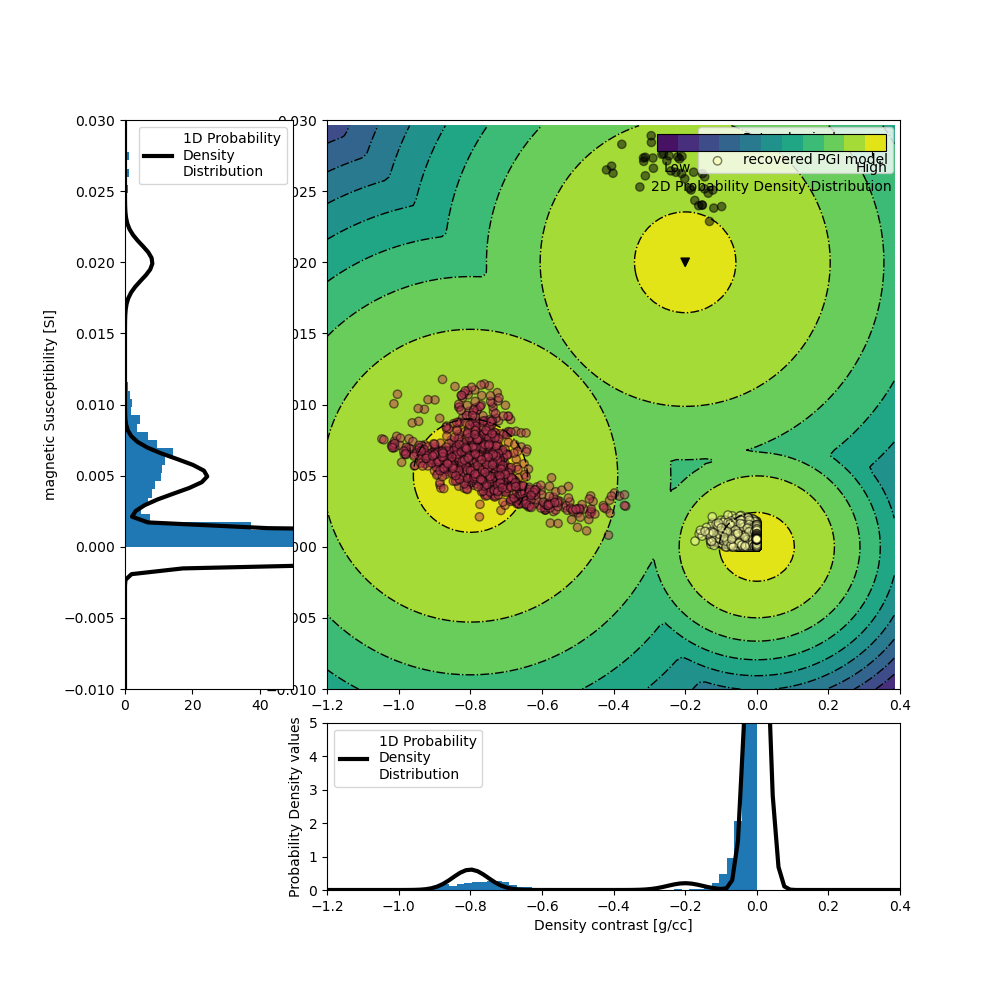
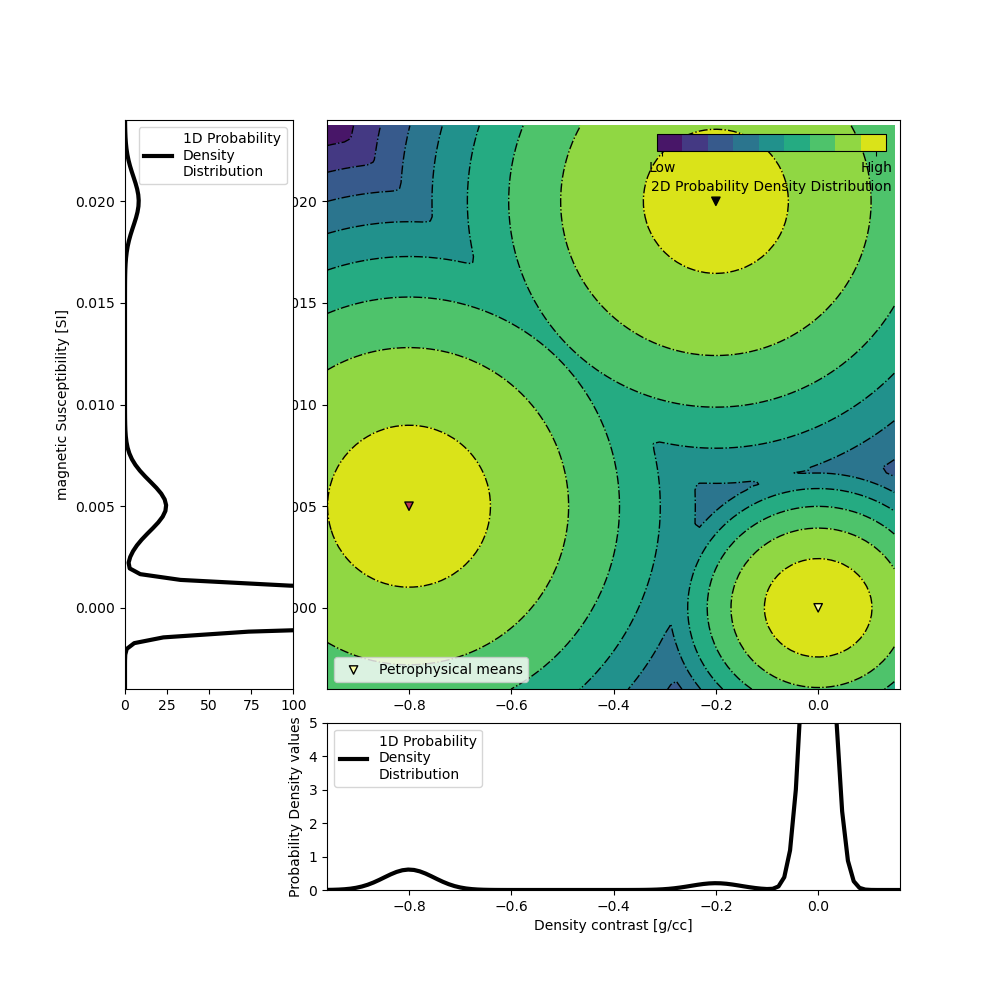
The GMM is our representation of the petrophysical and geological information. Here, we focus on the petrophysical aspect, with the means and covariances of the physical properties of each rock unit. To generate the data above, the PK unit was populated with a density contrast of -0.8 g/cc and a magnetic susceptibility of 0.005 SI. The properties of the HK unit were set at -0.2 g/cc and 0.02 SI. The covariances matrices are set so that we assume petrophysical noise levels of around 0.05 g/cc and 0.001 SI for both unit. Finally the background unit is set at null contrasts (0 g/cc 0 SI) with a petrophysical noise level of half of the above.
gmmref = utils.WeightedGaussianMixture(
n_components=3, # number of rock units: bckgrd, PK, HK
mesh=mesh, # inversion mesh
actv=actv, # actv cells
covariance_type="diag", # diagonal covariances
)
# required: initialization with fit
# fake random samples, size of the mesh, number of physical properties: 2 (density and mag.susc)
gmmref.fit(np.random.randn(nactv, 2))
# set parameters manually
# set phys. prop means for each unit
gmmref.means_ = np.c_[
[0.0, 0.0], # BCKGRD density contrast and mag. susc
[-0.8, 0.005], # PK
[-0.2, 0.02], # HK
].T
# set phys. prop covariances for each unit
gmmref.covariances_ = np.array(
[[6e-04, 3.175e-07], [2.4e-03, 1.5e-06], [2.4e-03, 1.5e-06]]
)
# important after setting cov. manually: compute precision matrices and cholesky
gmmref.compute_clusters_precisions()
# set global proportions; low-impact as long as not 0 or 1 (total=1)
gmmref.weights_ = np.r_[0.9, 0.075, 0.025]
# Plot the 2D GMM
ax = gmmref.plot_pdf(flag2d=True)
ax[0].set_xlabel("Density contrast [g/cc]")
ax[0].set_ylim([0, 5])
ax[2].set_ylabel("magnetic Susceptibility [SI]")
ax[2].set_xlim([0, 100])
plt.show()
Create PGI regularization#
# Sensitivity weighting
wr_grav = np.sum(simulation_grav.G**2.0, axis=0) ** 0.5 / (mesh.cell_volumes[actv])
wr_grav = wr_grav / np.max(wr_grav)
wr_mag = np.sum(simulation_mag.G**2.0, axis=0) ** 0.5 / (mesh.cell_volumes[actv])
wr_mag = wr_mag / np.max(wr_mag)
# create joint PGI regularization with smoothness
reg = regularization.PGI(
gmmref=gmmref,
mesh=mesh,
wiresmap=wires,
maplist=[idenMap, idenMap],
active_cells=actv,
alpha_pgi=1.0,
alpha_x=1.0,
alpha_y=1.0,
alpha_z=1.0,
alpha_xx=0.0,
alpha_yy=0.0,
alpha_zz=0.0,
# use the classification of the initial model (here, all background unit)
# as initial reference model
reference_model=utils.mkvc(
gmmref.means_[gmmref.predict(m0.reshape(actvMap.nP, -1))]
),
weights_list=[wr_grav, wr_mag], # weights each phys. prop. by correct sensW
)
Inverse problem with full petrophysical information#
# Directives
# Add directives to the inversion
# ratio to use for each phys prop. smoothness in each direction;
# roughly the ratio of the order of magnitude of each phys. prop.
alpha0_ratio = np.r_[
1e-4 * np.ones(len(reg.objfcts[1].objfcts[1:])),
1e-4 * 100.0 * np.ones(len(reg.objfcts[2].objfcts[1:])),
]
Alphas = directives.AlphasSmoothEstimate_ByEig(alpha0_ratio=alpha0_ratio, verbose=True)
# initialize beta and beta/alpha_pgi schedule
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-2)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
tolerance=0.2,
progress=0.2,
)
# geophy. and petro. target misfits
targets = directives.MultiTargetMisfits(
verbose=True,
)
# add learned mref in smooth once stable
MrefInSmooth = directives.PGI_AddMrefInSmooth(
wait_till_stable=True,
verbose=True,
)
# update the parameters in smallness (L2-approx of PGI)
update_smallness = directives.PGI_UpdateParameters(
update_gmm=False # keep GMM model fixed
)
# pre-conditioner
update_Jacobi = directives.UpdatePreconditioner()
# iteratively balance the scaling of the data misfits
scaling_init = directives.ScalingMultipleDataMisfits_ByEig(chi0_ratio=[1.0, 100.0])
scale_schedule = directives.JointScalingSchedule(verbose=True)
# Create inverse problem
# Optimization
# set lower and upper bounds
lowerbound = np.r_[-2.0 * np.ones(actvMap.nP), 0.0 * np.ones(actvMap.nP)]
upperbound = np.r_[0.0 * np.ones(actvMap.nP), 1e-1 * np.ones(actvMap.nP)]
opt = optimization.ProjectedGNCG(
maxIter=30,
lower=lowerbound,
upper=upperbound,
maxIterLS=20,
maxIterCG=100,
tolCG=1e-4,
)
# create inverse problem
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
inv = inversion.BaseInversion(
invProb,
# directives: evaluate alphas (and data misfits scales) before beta
directiveList=[
Alphas,
scaling_init,
beta,
update_smallness,
targets,
scale_schedule,
betaIt,
MrefInSmooth,
update_Jacobi,
],
)
# invert
pgi_model = inv.run(m0)
# Extract the results
density_model = gravmap * pgi_model
magsus_model = magmap * pgi_model
quasi_geology_model = actvMap * reg.objfcts[0].compute_quasi_geology_model()
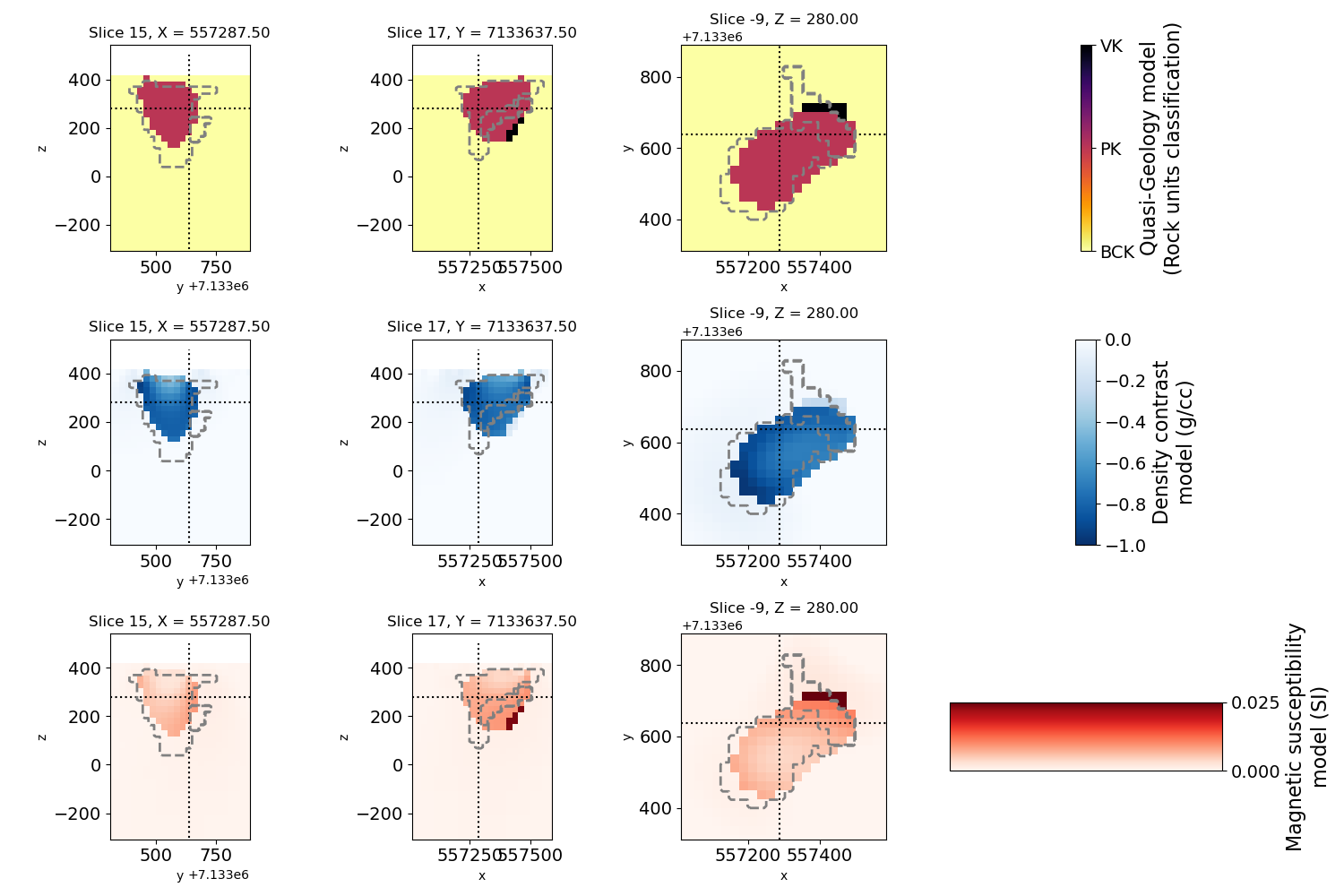
# Plot the result with full petrophysical information
fig, ax = plt.subplots(3, 4, figsize=(15, 10))
for _, axx in enumerate(ax):
for _, axxx in enumerate(axx):
axxx.set_aspect(1)
axxx.tick_params(labelsize=ticksize)
indx = 15
indy = 17
indz = -9
# geology model
mesh.plot_slice(
quasi_geology_model,
normal="X",
ax=ax[0, 0],
clim=[0, 2],
ind=indx,
pcolor_opts={"cmap": "inferno_r"},
)
mesh.plot_slice(
quasi_geology_model,
normal="Y",
ax=ax[0, 1],
clim=[0, 2],
ind=indy,
pcolor_opts={"cmap": "inferno_r"},
)
geoplot = mesh.plot_slice(
quasi_geology_model,
normal="Z",
ax=ax[0, 2],
clim=[0, 2],
ind=indz,
pcolor_opts={"cmap": "inferno_r"},
)
geocb = plt.colorbar(geoplot[0], cax=ax[0, 3], ticks=[0, 1, 2])
geocb.set_ticklabels(["BCK", "PK", "VK"])
geocb.set_label("Quasi-Geology model\n(Rock units classification)", fontsize=16)
ax[0, 3].set_aspect(10)
# gravity model
mesh.plot_slice(
density_model,
normal="X",
ax=ax[1, 0],
clim=[-1, 0],
ind=indx,
pcolor_opts={"cmap": "Blues_r"},
)
mesh.plot_slice(
density_model,
normal="Y",
ax=ax[1, 1],
clim=[-1, 0],
ind=indy,
pcolor_opts={"cmap": "Blues_r"},
)
denplot = mesh.plot_slice(
density_model,
normal="Z",
ax=ax[1, 2],
clim=[-1, 0],
ind=indz,
pcolor_opts={"cmap": "Blues_r"},
)
dencb = plt.colorbar(denplot[0], cax=ax[1, 3])
dencb.set_label("Density contrast\nmodel (g/cc)", fontsize=16)
ax[1, 3].set_aspect(10)
# magnetic model
mesh.plot_slice(
magsus_model,
normal="X",
ax=ax[2, 0],
clim=[0, 0.025],
ind=indx,
pcolor_opts={"cmap": "Reds"},
)
mesh.plot_slice(
magsus_model,
normal="Y",
ax=ax[2, 1],
clim=[0, 0.025],
ind=indy,
pcolor_opts={"cmap": "Reds"},
)
susplot = mesh.plot_slice(
magsus_model,
normal="Z",
ax=ax[2, 2],
clim=[0, 0.025],
ind=indz,
pcolor_opts={"cmap": "Reds"},
)
suscb = plt.colorbar(susplot[0], cax=ax[2, 3])
suscb.set_label("Magnetic susceptibility\nmodel (SI)", fontsize=16)
ax[2, 3].set_aspect(10)
# overlay true geology model for comparison
indslicexplot = mesh.gridCC[:, 0] == mesh.cell_centers_x[indx]
indsliceyplot = mesh.gridCC[:, 1] == mesh.cell_centers_y[indy]
indslicezplot = mesh.gridCC[:, 2] == mesh.cell_centers_z[indz]
for i in range(3):
for j, (plane, indd) in enumerate(
zip([[1, 2], [0, 2], [0, 1]], [indslicexplot, indsliceyplot, indslicezplot])
):
utils.plot2Ddata(
mesh.gridCC[indd][:, plane],
true_geology[indd],
nx=100,
ny=100,
contourOpts={"alpha": 0},
clim=[0, 2],
ax=ax[i, j],
level=True,
ncontour=2,
levelOpts={"colors": "grey", "linewidths": 2, "linestyles": "--"},
method="nearest",
)
# plot the locations of the cross-sections
for i in range(3):
ax[i, 0].plot(
mesh.cell_centers_y[indy] * np.ones(2), [-300, 500], c="k", linestyle="dotted"
)
ax[i, 0].plot(
[
data_mag.survey.receiver_locations[:, 1].min(),
data_mag.survey.receiver_locations[:, 1].max(),
],
mesh.cell_centers_z[indz] * np.ones(2),
c="k",
linestyle="dotted",
)
ax[i, 0].set_xlim(
[
data_mag.survey.receiver_locations[:, 1].min(),
data_mag.survey.receiver_locations[:, 1].max(),
],
)
ax[i, 1].plot(
mesh.cell_centers_x[indx] * np.ones(2), [-300, 500], c="k", linestyle="dotted"
)
ax[i, 1].plot(
[
data_mag.survey.receiver_locations[:, 0].min(),
data_mag.survey.receiver_locations[:, 0].max(),
],
mesh.cell_centers_z[indz] * np.ones(2),
c="k",
linestyle="dotted",
)
ax[i, 1].set_xlim(
[
data_mag.survey.receiver_locations[:, 0].min(),
data_mag.survey.receiver_locations[:, 0].max(),
],
)
ax[i, 2].plot(
mesh.cell_centers_x[indx] * np.ones(2),
[
data_mag.survey.receiver_locations[:, 1].min(),
data_mag.survey.receiver_locations[:, 1].max(),
],
c="k",
linestyle="dotted",
)
ax[i, 2].plot(
[
data_mag.survey.receiver_locations[:, 0].min(),
data_mag.survey.receiver_locations[:, 0].max(),
],
mesh.cell_centers_y[indy] * np.ones(2),
c="k",
linestyle="dotted",
)
ax[i, 2].set_xlim(
[
data_mag.survey.receiver_locations[:, 0].min(),
data_mag.survey.receiver_locations[:, 0].max(),
],
)
ax[i, 2].set_ylim(
[
data_mag.survey.receiver_locations[:, 1].min(),
data_mag.survey.receiver_locations[:, 1].max(),
],
)
plt.tight_layout()
plt.show()
# Plot the 2D GMM
fig = plt.figure(figsize=(10, 10))
ax0 = plt.subplot2grid((4, 4), (3, 1), colspan=3)
ax1 = plt.subplot2grid((4, 4), (0, 1), colspan=3, rowspan=3)
ax2 = plt.subplot2grid((4, 4), (0, 0), rowspan=3)
ax = [ax0, ax1, ax2]
reg.objfcts[0].gmm.plot_pdf(flag2d=True, ax=ax, padding=0.5)
ax[0].set_xlabel("Density contrast [g/cc]")
ax[0].set_ylim([0, 5])
ax[2].set_xlim([0, 50])
ax[2].set_ylabel("magnetic Susceptibility [SI]")
ax[1].scatter(
density_model[actv],
magsus_model[actv],
c=quasi_geology_model[actv],
cmap="inferno_r",
edgecolors="k",
label="recovered PGI model",
alpha=0.5,
)
ax[1].legend()
ax[0].hist(density_model[actv], density=True, bins=50)
ax[2].hist(magsus_model[actv], density=True, bins=50, orientation="horizontal")
plt.show()
Running inversion with SimPEG v0.22.2
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [83056.34689666449, 0.0, 62619.113223629334, 0.0, 132617.36938790858, 0.0, 10992873.22004899, 0.0, 7408640.753158514, 0.0, 20217611.386366405, 0.0]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.97141843 0.02858157]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 7.45e-06 4.23e+06 9.99e+04 4.23e+06 2.17e+02 0
geophys. misfits: 241593.2 (target 576.0 [False]); 46639.7 (target 576.0 [False]) | smallness misfit: 314552.8 (target: 23438.0 [False])
Beta cooling evaluation: progress: [241593.2 46639.7]; minimum progress targets: [3470231.3 572697.8]
mref changed in 794 places
1 7.45e-06 2.36e+05 3.10e+08 2.38e+05 1.41e+01 0
geophys. misfits: 6751.1 (target 576.0 [False]); 911.9 (target 576.0 [False]) | smallness misfit: 55948.2 (target: 23438.0 [False])
Beta cooling evaluation: progress: [6751.1 911.9]; minimum progress targets: [193274.6 37311.8]
mref changed in 24 places
2 7.45e-06 6.58e+03 1.36e+08 7.60e+03 2.07e+01 0
geophys. misfits: 3578.8 (target 576.0 [False]); 250.5 (target 576.0 [True]) | smallness misfit: 52393.8 (target: 23438.0 [False])
Updating scaling for data misfits by 2.2995260985909582
New scales: [0.98736661 0.01263339]
Beta cooling evaluation: progress: [3578.8 250.5]; minimum progress targets: [5400.9 729.6]
mref changed in 1 places
3 7.45e-06 3.54e+03 1.28e+08 4.49e+03 2.10e+01 0 Skip BFGS
geophys. misfits: 810.4 (target 576.0 [False]); 284.5 (target 576.0 [True]) | smallness misfit: 48854.0 (target: 23438.0 [False])
Updating scaling for data misfits by 2.0246576409225563
New scales: [0.99372008 0.00627992]
Beta cooling evaluation: progress: [810.4 284.5]; minimum progress targets: [2863. 691.2]
mref changed in 2 places
4 7.45e-06 8.07e+02 1.30e+08 1.77e+03 2.13e+01 0 Skip BFGS
geophys. misfits: 491.4 (target 576.0 [True]); 871.5 (target 576.0 [False]) | smallness misfit: 48729.2 (target: 23438.0 [False])
Updating scaling for data misfits by 1.1722612617795083
New scales: [0.99264625 0.00735375]
Beta cooling evaluation: progress: [491.4 871.5]; minimum progress targets: [691.2 691.2]
Decreasing beta to counter data misfit decrase plateau.
mref changed in 4 places
5 3.73e-06 4.94e+02 1.30e+08 9.77e+02 3.75e+01 0 Skip BFGS
geophys. misfits: 435.2 (target 576.0 [True]); 350.1 (target 576.0 [True]) | smallness misfit: 51016.9 (target: 23438.0 [False])
Beta cooling evaluation: progress: [435.2 350.1]; minimum progress targets: [691.2 697.2]
Warming alpha_pgi to favor clustering: 1.4843795575722702
mref changed in 11 places
6 3.73e-06 4.35e+02 1.83e+08 1.12e+03 3.19e+01 0
geophys. misfits: 379.1 (target 576.0 [True]); 330.8 (target 576.0 [True]) | smallness misfit: 38637.3 (target: 23438.0 [False])
Beta cooling evaluation: progress: [379.1 330.8]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 2.4200594420540003
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
7 3.73e-06 3.79e+02 2.38e+08 1.27e+03 2.27e+01 0
geophys. misfits: 409.6 (target 576.0 [True]); 887.4 (target 576.0 [False]) | smallness misfit: 33909.4 (target: 23438.0 [False])
Updating scaling for data misfits by 1.4060930485367058
New scales: [0.98969073 0.01030927]
Beta cooling evaluation: progress: [409.6 887.4]; minimum progress targets: [691.2 691.2]
Decreasing beta to counter data misfit increase.
mref changed in 0 places
8 1.86e-06 4.15e+02 2.20e+08 8.24e+02 4.48e+01 0
geophys. misfits: 330.0 (target 576.0 [True]); 296.4 (target 576.0 [True]) | smallness misfit: 36049.7 (target: 23438.0 [False])
Beta cooling evaluation: progress: [330. 296.4]; minimum progress targets: [691.2 709.9]
Warming alpha_pgi to favor clustering: 4.463534518304005
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
9 1.86e-06 3.30e+02 3.97e+08 1.07e+03 2.38e+01 0
geophys. misfits: 386.0 (target 576.0 [True]); 260.0 (target 576.0 [True]) | smallness misfit: 29857.1 (target: 23438.0 [False])
Beta cooling evaluation: progress: [386. 260.]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 8.274660214090309
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
10 1.86e-06 3.85e+02 5.63e+08 1.43e+03 2.34e+01 0
geophys. misfits: 484.1 (target 576.0 [True]); 510.5 (target 576.0 [True]) | smallness misfit: 27007.8 (target: 23438.0 [False])
Beta cooling evaluation: progress: [484.1 510.5]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 9.59089181250864
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
11 1.86e-06 4.84e+02 5.53e+08 1.52e+03 2.34e+01 0
geophys. misfits: 494.8 (target 576.0 [True]); 575.0 (target 576.0 [True]) | smallness misfit: 27114.4 (target: 23438.0 [False])
Beta cooling evaluation: progress: [494.8 575. ]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 10.386108294220053
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
12 1.86e-06 4.96e+02 5.81e+08 1.58e+03 3.67e+01 0 Skip BFGS
geophys. misfits: 507.6 (target 576.0 [True]); 671.1 (target 576.0 [False]) | smallness misfit: 26900.2 (target: 23438.0 [False])
Updating scaling for data misfits by 1.1346429603742938
New scales: [0.98831888 0.01168112]
Beta cooling evaluation: progress: [507.6 671.1]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
13 1.86e-06 5.10e+02 5.73e+08 1.58e+03 4.23e+01 0
geophys. misfits: 518.2 (target 576.0 [True]); 483.5 (target 576.0 [True]) | smallness misfit: 25534.2 (target: 23438.0 [False])
Beta cooling evaluation: progress: [518.2 483.5]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 11.959545141301799
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
14 1.86e-06 5.18e+02 6.35e+08 1.70e+03 2.30e+01 0
geophys. misfits: 526.3 (target 576.0 [True]); 493.0 (target 576.0 [True]) | smallness misfit: 25439.3 (target: 23438.0 [False])
Beta cooling evaluation: progress: [526.3 493. ]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.530713816908548
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
15 1.86e-06 5.26e+02 7.00e+08 1.83e+03 2.29e+01 2 Skip BFGS
geophys. misfits: 570.8 (target 576.0 [True]); 659.8 (target 576.0 [False]) | smallness misfit: 25022.8 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0090477116022927
New scales: [0.98821443 0.01178557]
Beta cooling evaluation: progress: [570.8 659.8]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
16 1.86e-06 5.72e+02 6.70e+08 1.82e+03 1.34e+01 0
geophys. misfits: 570.5 (target 576.0 [True]); 590.1 (target 576.0 [False]) | smallness misfit: 24805.9 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0095909089963857
New scales: [0.98810274 0.01189726]
Beta cooling evaluation: progress: [570.5 590.1]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
17 1.86e-06 5.71e+02 6.70e+08 1.82e+03 5.87e+00 0
geophys. misfits: 570.5 (target 576.0 [True]); 592.2 (target 576.0 [False]) | smallness misfit: 24787.9 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0095641373782287
New scales: [0.98799032 0.01200968]
Beta cooling evaluation: progress: [570.5 592.2]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
18 1.86e-06 5.71e+02 6.70e+08 1.82e+03 9.16e+00 3 Skip BFGS
geophys. misfits: 570.7 (target 576.0 [True]); 582.5 (target 576.0 [False]) | smallness misfit: 24842.6 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0092800404005149
New scales: [0.98788022 0.01211978]
Beta cooling evaluation: progress: [570.7 582.5]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
19 1.86e-06 5.71e+02 6.70e+08 1.82e+03 5.84e+00 0
geophys. misfits: 570.7 (target 576.0 [True]); 577.7 (target 576.0 [False]) | smallness misfit: 24843.5 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0092488213843107
New scales: [0.9877695 0.0122305]
Beta cooling evaluation: progress: [570.7 577.7]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
20 1.86e-06 5.71e+02 6.70e+08 1.82e+03 3.17e+00 0
geophys. misfits: 570.7 (target 576.0 [True]); 572.0 (target 576.0 [True]) | smallness misfit: 24849.0 (target: 23438.0 [False])
Beta cooling evaluation: progress: [570.7 572. ]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.640942187948006
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
21 1.86e-06 5.71e+02 6.75e+08 1.83e+03 3.05e+00 0
geophys. misfits: 572.6 (target 576.0 [True]); 577.2 (target 576.0 [False]) | smallness misfit: 24821.7 (target: 23438.0 [False])
Updating scaling for data misfits by 1.005905737800258
New scales: [0.98769816 0.01230184]
Beta cooling evaluation: progress: [572.6 577.2]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
22 1.86e-06 5.73e+02 6.74e+08 1.83e+03 5.04e+00 0
geophys. misfits: 572.6 (target 576.0 [True]); 573.8 (target 576.0 [True]) | smallness misfit: 24826.8 (target: 23438.0 [False])
Beta cooling evaluation: progress: [572.6 573.8]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.707587673092597
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
23 1.86e-06 5.73e+02 6.77e+08 1.83e+03 2.43e+00 0
geophys. misfits: 573.8 (target 576.0 [True]); 576.8 (target 576.0 [False]) | smallness misfit: 24812.7 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0038983791657345
New scales: [0.98765079 0.01234921]
Beta cooling evaluation: progress: [573.8 576.8]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
24 1.86e-06 5.74e+02 6.76e+08 1.83e+03 3.03e+00 0
geophys. misfits: 573.8 (target 576.0 [True]); 575.7 (target 576.0 [True]) | smallness misfit: 24813.8 (target: 23438.0 [False])
Beta cooling evaluation: progress: [573.8 575.7]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.737835943349753
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
25 1.86e-06 5.74e+02 6.77e+08 1.84e+03 1.58e+00 1
geophys. misfits: 574.3 (target 576.0 [True]); 576.0 (target 576.0 [False]) | smallness misfit: 24808.7 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0029875471003311
New scales: [0.98761436 0.01238564]
Beta cooling evaluation: progress: [574.3 576. ]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
26 1.86e-06 5.74e+02 6.77e+08 1.84e+03 2.92e+00 0
geophys. misfits: 574.3 (target 576.0 [True]); 574.2 (target 576.0 [True]) | smallness misfit: 24810.2 (target: 23438.0 [False])
Beta cooling evaluation: progress: [574.3 574.2]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.779862809400854
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
27 1.86e-06 5.74e+02 6.79e+08 1.84e+03 1.99e+00 0
geophys. misfits: 575.0 (target 576.0 [True]); 575.9 (target 576.0 [True]) | smallness misfit: 24801.4 (target: 23438.0 [False])
Beta cooling evaluation: progress: [575. 575.9]; minimum progress targets: [691.2 691.2]
Warming alpha_pgi to favor clustering: 13.7927390493134
mref changed in 0 places
Add mref to Smoothness. Changes in mref happened in 0.0 % of the cells
28 1.86e-06 5.75e+02 6.79e+08 1.84e+03 9.48e-01 0
geophys. misfits: 575.2 (target 576.0 [True]); 576.7 (target 576.0 [False]) | smallness misfit: 24798.8 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0013520305076358
New scales: [0.98759782 0.01240218]
Beta cooling evaluation: progress: [575.2 576.7]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
29 1.86e-06 5.75e+02 6.79e+08 1.84e+03 2.15e+00 0 Skip BFGS
geophys. misfits: 575.2 (target 576.0 [True]); 576.7 (target 576.0 [False]) | smallness misfit: 24798.8 (target: 23438.0 [False])
Updating scaling for data misfits by 1.0013520305076358
New scales: [0.98758126 0.01241874]
Beta cooling evaluation: progress: [575.2 576.7]; minimum progress targets: [691.2 691.2]
mref changed in 0 places
30 1.86e-06 5.75e+02 6.79e+08 1.84e+03 2.81e+00 18
------------------------- STOP! -------------------------
1 : |fc-fOld| = 2.3930e-05 <= tolF*(1+|f0|) = 4.2343e+05
1 : |xc-x_last| = 1.0548e-10 <= tolX*(1+|x0|) = 1.0153e-01
0 : |proj(x-g)-x| = 2.8068e+00 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 2.8068e+00 <= 1e3*eps = 1.0000e-02
1 : maxIter = 30 <= iter = 30
------------------------- DONE! -------------------------
Total running time of the script: (4 minutes 14.280 seconds)
Estimated memory usage: 246 MB