Note
Go to the end to download the full example code
Petrophysically guided inversion: Joint linear example with nonlinear relationships#
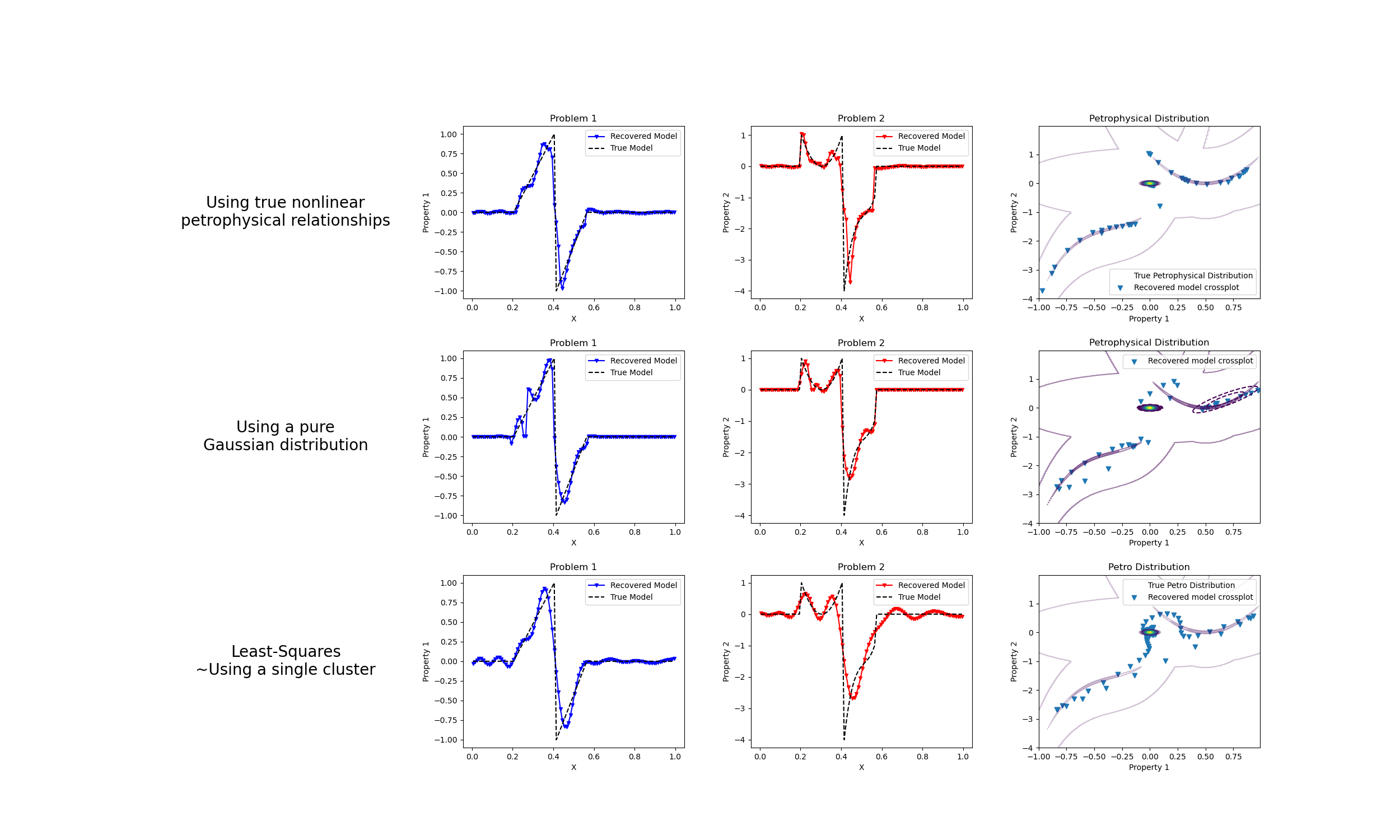
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically guided inversion. We explore it through coupling two linear problems whose respective physical properties are linked by polynomial relationships that change between rock units.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.466585984471504, 0.0, 3.4976674770427723e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09601038 0.90398962]
<class 'SimPEG.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09601038 0.90398962]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.92e+01 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 1059.2 (target 30.0 [False]); 83.6 (target 30.0 [False]) | smallness misfit: 2946.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [1059.2 83.6]; minimum progress targets: [240000. 240000.]
1 1.92e+01 1.77e+02 4.12e+01 9.69e+02 8.94e+01 0
geophys. misfits: 482.6 (target 30.0 [False]); 29.2 (target 30.0 [True]) | smallness misfit: 1332.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [482.6 29.2]; minimum progress targets: [847.4 66.9]
Updating scaling for data misfits by 1.0270003248200115
New scales: [0.09834774 0.90165226]
2 1.92e+01 7.38e+01 4.00e+01 8.43e+02 7.76e+01 0 Skip BFGS
geophys. misfits: 459.7 (target 30.0 [False]); 29.1 (target 30.0 [True]) | smallness misfit: 1312.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [459.7 29.1]; minimum progress targets: [386.1 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.0314593904410994
New scales: [0.10112882 0.89887118]
3 9.60e+00 7.26e+01 4.01e+01 4.58e+02 8.40e+01 0 Skip BFGS
geophys. misfits: 167.3 (target 30.0 [False]); 24.6 (target 30.0 [True]) | smallness misfit: 1317.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [167.3 24.6]; minimum progress targets: [367.7 30. ]
Updating scaling for data misfits by 1.219948708502077
New scales: [0.1206875 0.8793125]
4 9.60e+00 4.18e+01 4.25e+01 4.50e+02 7.39e+01 0
geophys. misfits: 143.5 (target 30.0 [False]); 25.0 (target 30.0 [True]) | smallness misfit: 1265.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [143.5 25. ]; minimum progress targets: [133.8 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.1996666333296457
New scales: [0.14137794 0.85862206]
5 4.80e+00 4.18e+01 4.27e+01 2.47e+02 7.75e+01 0 Skip BFGS
geophys. misfits: 57.8 (target 30.0 [False]); 22.5 (target 30.0 [True]) | smallness misfit: 1372.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [57.8 22.5]; minimum progress targets: [114.8 30. ]
Updating scaling for data misfits by 1.3342957626919507
New scales: [0.18012683 0.81987317]
6 4.80e+00 2.89e+01 4.46e+01 2.43e+02 7.44e+01 0
geophys. misfits: 43.0 (target 30.0 [False]); 22.4 (target 30.0 [True]) | smallness misfit: 1309.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [43. 22.4]; minimum progress targets: [46.3 30. ]
Updating scaling for data misfits by 1.3375229024814719
New scales: [0.22711581 0.77288419]
7 4.80e+00 2.71e+01 4.48e+01 2.42e+02 7.33e+01 0
geophys. misfits: 32.2 (target 30.0 [False]); 22.4 (target 30.0 [True]) | smallness misfit: 1276.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [32.2 22.4]; minimum progress targets: [34.4 30. ]
Updating scaling for data misfits by 1.3420194632227151
New scales: [0.28282458 0.71717542]
8 4.80e+00 2.51e+01 4.52e+01 2.42e+02 7.54e+01 0
geophys. misfits: 26.0 (target 30.0 [True]); 22.4 (target 30.0 [True]) | smallness misfit: 1250.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [26. 22.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.2449219097000142
9 4.80e+00 2.34e+01 4.61e+01 2.45e+02 6.14e+01 0
geophys. misfits: 25.3 (target 30.0 [True]); 22.8 (target 30.0 [True]) | smallness misfit: 1187.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.3 22.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.5573370442568115
10 4.80e+00 2.35e+01 4.69e+01 2.49e+02 4.67e+01 0
geophys. misfits: 24.3 (target 30.0 [True]); 23.4 (target 30.0 [True]) | smallness misfit: 1106.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.3 23.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.95922803198821
11 4.80e+00 2.36e+01 4.78e+01 2.53e+02 3.48e+01 0
geophys. misfits: 23.6 (target 30.0 [True]); 24.0 (target 30.0 [True]) | smallness misfit: 1025.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.6 24. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.467445418444921
12 4.80e+00 2.39e+01 4.88e+01 2.58e+02 3.67e+01 0
geophys. misfits: 23.7 (target 30.0 [True]); 24.7 (target 30.0 [True]) | smallness misfit: 953.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.7 24.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.0632447674595578
13 4.80e+00 2.44e+01 4.99e+01 2.64e+02 6.00e+01 0
geophys. misfits: 22.7 (target 30.0 [True]); 25.4 (target 30.0 [True]) | smallness misfit: 880.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.7 25.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.8375817995772943
14 4.80e+00 2.46e+01 5.13e+01 2.71e+02 7.30e+01 0
geophys. misfits: 22.0 (target 30.0 [True]); 26.1 (target 30.0 [True]) | smallness misfit: 804.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [22. 26.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 4.821066325745199
15 4.80e+00 2.49e+01 5.29e+01 2.79e+02 8.00e+01 0
geophys. misfits: 22.7 (target 30.0 [True]); 27.1 (target 30.0 [True]) | smallness misfit: 733.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.7 27.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 5.860579033410637
16 4.80e+00 2.58e+01 5.43e+01 2.87e+02 8.09e+01 0
geophys. misfits: 22.2 (target 30.0 [True]); 27.8 (target 30.0 [True]) | smallness misfit: 678.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.2 27.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 7.1189553468605435
17 4.80e+00 2.62e+01 5.61e+01 2.96e+02 8.29e+01 0
geophys. misfits: 22.2 (target 30.0 [True]); 28.5 (target 30.0 [True]) | smallness misfit: 615.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.2 28.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 8.541098513892216
18 4.80e+00 2.68e+01 5.78e+01 3.04e+02 9.42e+01 0
geophys. misfits: 23.5 (target 30.0 [True]); 29.9 (target 30.0 [True]) | smallness misfit: 562.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.5 29.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 9.737412991924352
19 4.80e+00 2.81e+01 5.89e+01 3.11e+02 8.32e+01 0
geophys. misfits: 25.7 (target 30.0 [True]); 30.7 (target 30.0 [False]) | smallness misfit: 514.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.7 30.7]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.1690573002451738
New scales: [0.25224184 0.74775816]
20 2.40e+00 2.94e+01 5.85e+01 1.70e+02 9.94e+01 0
geophys. misfits: 19.6 (target 30.0 [True]); 25.8 (target 30.0 [True]) | smallness misfit: 543.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [19.6 25.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 13.08986157124665
21 2.40e+00 2.43e+01 6.40e+01 1.78e+02 8.59e+01 0
geophys. misfits: 18.4 (target 30.0 [True]); 25.5 (target 30.0 [True]) | smallness misfit: 490.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [18.4 25.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 18.363780573348887
22 2.40e+00 2.37e+01 6.98e+01 1.91e+02 1.01e+02 0
geophys. misfits: 17.6 (target 30.0 [True]); 24.7 (target 30.0 [True]) | smallness misfit: 442.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.6 24.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 26.851793413284888
23 2.40e+00 2.29e+01 7.80e+01 2.10e+02 1.08e+02 0
geophys. misfits: 17.3 (target 30.0 [True]); 23.8 (target 30.0 [True]) | smallness misfit: 391.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.3 23.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 40.17971519844774
24 2.40e+00 2.22e+01 8.94e+01 2.37e+02 1.15e+02 0
geophys. misfits: 17.3 (target 30.0 [True]); 24.3 (target 30.0 [True]) | smallness misfit: 345.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.3 24.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 59.741920800674485
25 2.40e+00 2.25e+01 1.03e+02 2.70e+02 1.18e+02 0
geophys. misfits: 21.1 (target 30.0 [True]); 23.0 (target 30.0 [True]) | smallness misfit: 301.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.1 23. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 81.35391606203183
26 2.40e+00 2.26e+01 1.16e+02 3.02e+02 1.19e+02 0
geophys. misfits: 22.9 (target 30.0 [True]); 24.4 (target 30.0 [True]) | smallness misfit: 291.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.9 24.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 103.18660220854213
27 2.40e+00 2.41e+01 1.29e+02 3.33e+02 1.22e+02 0
geophys. misfits: 25.2 (target 30.0 [True]); 23.9 (target 30.0 [True]) | smallness misfit: 225.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.2 23.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 126.07099988334166
28 2.40e+00 2.42e+01 1.37e+02 3.54e+02 1.24e+02 0
geophys. misfits: 39.6 (target 30.0 [False]); 28.4 (target 30.0 [True]) | smallness misfit: 193.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [39.6 28.4]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.055587985138429
New scales: [0.26258164 0.73741836]
29 1.20e+00 3.14e+01 1.28e+02 1.85e+02 1.23e+02 0
geophys. misfits: 26.1 (target 30.0 [True]); 28.3 (target 30.0 [True]) | smallness misfit: 206.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.1 28.3]; minimum progress targets: [31.7 30. ]
Warming alpha_pgi to favor clustering: 139.25290145897227
30 1.20e+00 2.77e+01 1.32e+02 1.86e+02 1.16e+02 0
geophys. misfits: 25.2 (target 30.0 [True]); 26.5 (target 30.0 [True]) | smallness misfit: 193.1 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [25.2 26.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 161.7783957915216
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 7.0273e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 1.1632e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.1632e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 31
------------------------- DONE! -------------------------
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [0.0003449902502617977, 0.0, 3.7949084087632503e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09601038 0.90398962]
<class 'SimPEG.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09601038 0.90398962]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.91e+03 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 87797.2 (target 30.0 [False]); 62285.6 (target 30.0 [False]) | smallness misfit: 275.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [87797.2 62285.6]; minimum progress targets: [240000. 240000.]
1 1.91e+03 6.47e+04 6.69e-01 6.60e+04 9.27e+01 0
geophys. misfits: 627.9 (target 30.0 [False]); 27.8 (target 30.0 [True]) | smallness misfit: 124.8 (target: 200.0 [True])
Beta cooling evaluation: progress: [627.9 27.8]; minimum progress targets: [70237.8 49828.5]
Updating scaling for data misfits by 1.0777978985257075
New scales: [0.10271258 0.89728742]
2 1.91e+03 8.95e+01 3.37e-01 7.34e+02 7.85e+01 0 Skip BFGS
geophys. misfits: 111.3 (target 30.0 [False]); 26.1 (target 30.0 [True]) | smallness misfit: 47.8 (target: 200.0 [True])
Beta cooling evaluation: progress: [111.3 26.1]; minimum progress targets: [502.3 30. ]
Updating scaling for data misfits by 1.149353459035614
New scales: [0.11626944 0.88373056]
3 1.91e+03 3.60e+01 1.32e-01 2.89e+02 8.30e+01 0 Skip BFGS
geophys. misfits: 98.0 (target 30.0 [False]); 24.5 (target 30.0 [True]) | smallness misfit: 51.5 (target: 200.0 [True])
Beta cooling evaluation: progress: [98. 24.5]; minimum progress targets: [89. 30.]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.2240032601152608
New scales: [0.13870172 0.86129828]
4 9.57e+02 3.47e+01 1.35e-01 1.64e+02 9.86e+01 0
geophys. misfits: 36.3 (target 30.0 [False]); 21.9 (target 30.0 [True]) | smallness misfit: 49.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [36.3 21.9]; minimum progress targets: [78.4 30. ]
Updating scaling for data misfits by 1.3684205634446134
New scales: [0.18057481 0.81942519]
5 9.57e+02 2.45e+01 1.33e-01 1.52e+02 5.30e+01 0
geophys. misfits: 28.9 (target 30.0 [True]); 22.1 (target 30.0 [True]) | smallness misfit: 49.7 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [28.9 22.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.1972365675938115
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 6.2925e-02 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 5.3045e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 5.3045e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 6
------------------------- DONE! -------------------------
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.5188574849327566e-05, 0.0, 3.4826374354602616e-05, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09601038 0.90398962]
/home/vsts/work/1/s/SimPEG/directives/directives.py:332: UserWarning:
There is no PGI regularization. Smallness target is turned off (TriggerSmall flag)
Initial data misfit scales: [0.09601038 0.90398962]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.04e+06 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 56377.6 (target 30.0 [False]); 35775.8 (target 30.0 [False])
1 2.08e+05 3.78e+04 4.29e-02 4.67e+04 1.38e+02 0
geophys. misfits: 8375.2 (target 30.0 [False]); 3857.2 (target 30.0 [False])
2 4.15e+04 4.29e+03 1.07e-01 8.73e+03 1.31e+02 0 Skip BFGS
geophys. misfits: 563.0 (target 30.0 [False]); 256.2 (target 30.0 [False])
3 8.30e+03 2.86e+02 1.41e-01 1.46e+03 1.01e+02 0 Skip BFGS
geophys. misfits: 42.6 (target 30.0 [False]); 38.2 (target 30.0 [False])
4 1.66e+03 3.86e+01 1.51e-01 2.90e+02 7.91e+01 0 Skip BFGS
geophys. misfits: 17.6 (target 30.0 [True]); 22.1 (target 30.0 [True])
All targets have been reached
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 4.8127e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 7.9019e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 7.9019e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:301: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:308: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:346: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:353: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:360: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:368: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:402: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:409: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
import discretize as Mesh
import matplotlib.pyplot as plt
import numpy as np
from SimPEG import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 30
jk = np.linspace(1.0, 59.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
m0 = np.zeros(mesh.nC)
m0[20:41] = np.linspace(0.0, 1.0, 21)
m0[41:57] = np.linspace(-1, 0.0, 16)
poly0 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[0.0, -4.0, 4.0])
poly1 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[-0.0, 3.0, 6.0, 6.0])
poly0_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, -4.0, 4.0])
poly1_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, 3.0, 6.0, 6.0])
cluster_mapping = [maps.IdentityMap(), poly0_inverse, poly1_inverse]
m1 = np.zeros(100)
m1[20:41] = 1.0 + (poly0 * np.vstack([m0[20:41], m1[20:41]]).T)[:, 1]
m1[41:57] = -1.0 + (poly1 * np.vstack([m0[41:57], m1[41:57]]).T)[:, 1]
model2d = np.vstack([m0, m1]).T
m = utils.mkvc(model2d)
clfmapping = utils.GaussianMixtureWithNonlinearRelationships(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
means_init=np.array(
[
[0, 0],
[m0[20:41].mean(), m1[20:41].mean()],
[m0[41:57].mean(), m1[41:57].mean()],
]
),
verbose=0,
verbose_interval=10,
cluster_mapping=cluster_mapping,
)
clfmapping = clfmapping.fit(model2d)
clfnomapping = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
verbose=0,
verbose_interval=10,
)
clfnomapping = clfnomapping.fit(model2d)
wires = maps.Wires(("m1", mesh.nC), ("m2", mesh.nC))
relatrive_error = 0.01
noise_floor = 0.0
prob1 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m1)
survey1 = prob1.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
prob2 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m2)
survey2 = prob2.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
dmis1 = data_misfit.L2DataMisfit(simulation=prob1, data=survey1)
dmis2 = data_misfit.L2DataMisfit(simulation=prob2, data=survey2)
dmis = dmis1 + dmis2
minit = np.zeros_like(m)
# Distance weighting
wr1 = np.sum(prob1.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr1 = wr1 / np.max(wr1)
wr2 = np.sum(prob2.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr2 = wr2 / np.max(wr2)
reg_simple = regularization.PGI(
mesh=mesh,
gmmref=clfmapping,
gmm=clfmapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=True,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[1e6, 1e4, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_map = inv.run(minit)
# Inversion with no nonlinear mapping
reg_simple_no_map = regularization.PGI(
mesh=mesh,
gmmref=clfnomapping,
gmm=clfnomapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=False,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple_no_map, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[100.0 * np.ones(2), 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(
chiSmall=1.0, TriggerSmall=True, TriggerTheta=False, verbose=True
)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_no_map = inv.run(minit)
# WeightedLeastSquares Inversion
reg1 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m1, weights={"cell_weights": wr1}
)
reg2 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m2, weights={"cell_weights": wr2}
)
reg = reg1 + reg2
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
alpha0_ratio = np.r_[1, 1, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
beta_schedule = directives.BetaSchedule(coolingFactor=5.0, coolingRate=1)
targets = directives.MultiTargetMisfits(
TriggerSmall=False,
verbose=True,
)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, targets, beta_schedule, scaling_schedule],
)
mtik = inv.run(minit)
# Final Plot
fig, axes = plt.subplots(3, 4, figsize=(25, 15))
axes = axes.reshape(12)
left, width = 0.25, 0.5
bottom, height = 0.25, 0.5
right = left + width
top = bottom + height
axes[0].set_axis_off()
axes[0].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using true nonlinear\npetrophysical relationships"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[0].transAxes,
)
axes[1].plot(mesh.cell_centers_x, wires.m1 * mcluster_map, "b.-", ms=5, marker="v")
axes[1].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[1].set_title("Problem 1")
axes[1].legend(["Recovered Model", "True Model"], loc=1)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Property 1")
axes[2].plot(mesh.cell_centers_x, wires.m2 * mcluster_map, "r.-", ms=5, marker="v")
axes[2].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[2].set_title("Problem 2")
axes[2].legend(["Recovered Model", "True Model"], loc=1)
axes[2].set_xlabel("X")
axes[2].set_ylabel("Property 2")
x, y = np.mgrid[-1:1:0.01, -4:2:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
CS = axes[3].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[3].scatter(wires.m1 * mcluster_map, wires.m2 * mcluster_map, marker="v")
axes[3].set_title("Petrophysical Distribution")
CS.collections[0].set_label("")
axes[3].legend(["True Petrophysical Distribution", "Recovered model crossplot"])
axes[3].set_xlabel("Property 1")
axes[3].set_ylabel("Property 2")
axes[4].set_axis_off()
axes[4].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using a pure\nGaussian distribution"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[4].transAxes,
)
axes[5].plot(mesh.cell_centers_x, wires.m1 * mcluster_no_map, "b.-", ms=5, marker="v")
axes[5].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[5].set_title("Problem 1")
axes[5].legend(["Recovered Model", "True Model"], loc=1)
axes[5].set_xlabel("X")
axes[5].set_ylabel("Property 1")
axes[6].plot(mesh.cell_centers_x, wires.m2 * mcluster_no_map, "r.-", ms=5, marker="v")
axes[6].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[6].set_title("Problem 2")
axes[6].legend(["Recovered Model", "True Model"], loc=1)
axes[6].set_xlabel("X")
axes[6].set_ylabel("Property 2")
CSF = axes[7].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.5,
label="True Petro. Distribution",
)
CS = axes[7].contour(
x,
y,
np.exp(clfnomapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
500,
cmap="viridis",
linestyles="--",
label="Modeled Petro. Distribution",
)
axes[7].scatter(
wires.m1 * mcluster_no_map,
wires.m2 * mcluster_no_map,
marker="v",
label="Recovered model crossplot",
)
axes[7].set_title("Petrophysical Distribution")
axes[7].legend()
axes[7].set_xlabel("Property 1")
axes[7].set_ylabel("Property 2")
# Tikonov
axes[8].set_axis_off()
axes[8].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Least-Squares\n~Using a single cluster"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[8].transAxes,
)
axes[9].plot(mesh.cell_centers_x, wires.m1 * mtik, "b.-", ms=5, marker="v")
axes[9].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[9].set_title("Problem 1")
axes[9].legend(["Recovered Model", "True Model"], loc=1)
axes[9].set_xlabel("X")
axes[9].set_ylabel("Property 1")
axes[10].plot(mesh.cell_centers_x, wires.m2 * mtik, "r.-", ms=5, marker="v")
axes[10].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[10].set_title("Problem 2")
axes[10].legend(["Recovered Model", "True Model"], loc=1)
axes[10].set_xlabel("X")
axes[10].set_ylabel("Property 2")
CS = axes[11].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[11].scatter(wires.m1 * mtik, wires.m2 * mtik, marker="v")
axes[11].set_title("Petro Distribution")
CS.collections[0].set_label("")
axes[11].legend(["True Petro Distribution", "Recovered model crossplot"])
axes[11].set_xlabel("Property 1")
axes[11].set_ylabel("Property 2")
plt.subplots_adjust(wspace=0.3, hspace=0.3, top=0.85)
plt.show()
Total running time of the script: (0 minutes 34.045 seconds)
Estimated memory usage: 8 MB