Note
Go to the end to download the full example code.
Petrophysically guided inversion: Joint linear example with nonlinear relationships#
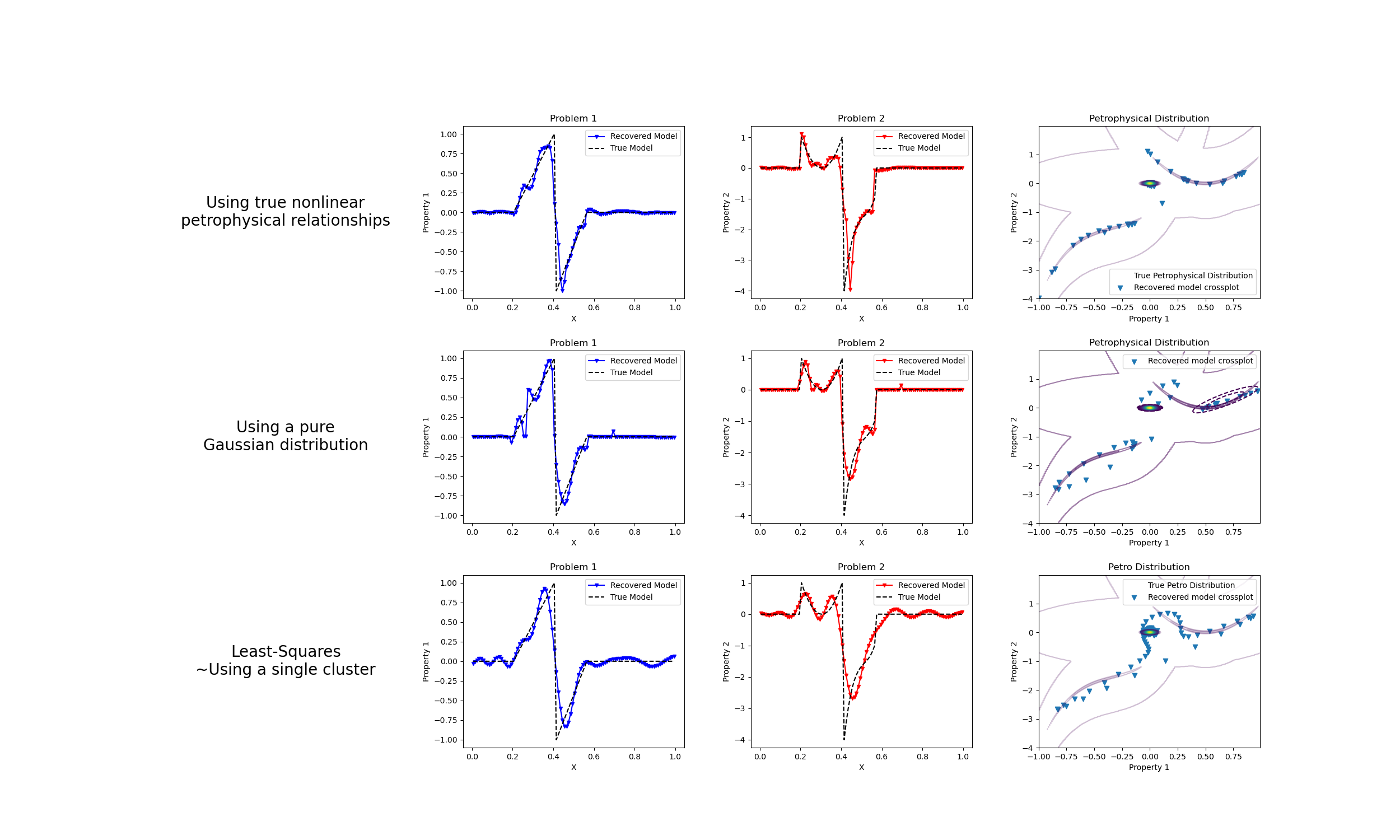
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically guided inversion. We explore it through coupling two linear problems whose respective physical properties are linked by polynomial relationships that change between rock units.
Running inversion with SimPEG v0.22.1
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.49503918967184, 0.0, 3.6131602071908994e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09850237 0.90149763]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09850237 0.90149763]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.95e+01 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 1037.7 (target 30.0 [False]); 81.3 (target 30.0 [False]) | smallness misfit: 2942.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [1037.7 81.3]; minimum progress targets: [240000. 240000.]
1 1.95e+01 1.75e+02 4.15e+01 9.86e+02 8.68e+01 0
geophys. misfits: 474.9 (target 30.0 [False]); 25.3 (target 30.0 [True]) | smallness misfit: 1334.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [474.9 25.3]; minimum progress targets: [830.2 65. ]
Updating scaling for data misfits by 1.1861041074440497
New scales: [0.11473085 0.88526915]
2 1.95e+01 7.69e+01 4.02e+01 8.63e+02 6.07e+01 0 Skip BFGS
geophys. misfits: 377.2 (target 30.0 [False]); 25.1 (target 30.0 [True]) | smallness misfit: 1288.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [377.2 25.1]; minimum progress targets: [379.9 30. ]
Updating scaling for data misfits by 1.1947439955698549
New scales: [0.13407827 0.86592173]
3 1.95e+01 7.23e+01 4.07e+01 8.69e+02 6.61e+01 0 Skip BFGS
geophys. misfits: 300.7 (target 30.0 [False]); 25.1 (target 30.0 [True]) | smallness misfit: 1252.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [300.7 25.1]; minimum progress targets: [301.7 30. ]
Updating scaling for data misfits by 1.197372183297028
New scales: [0.15640266 0.84359734]
4 1.95e+01 6.82e+01 4.12e+01 8.74e+02 5.85e+01 0 Skip BFGS
geophys. misfits: 240.9 (target 30.0 [False]); 25.1 (target 30.0 [True]) | smallness misfit: 1224.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [240.9 25.1]; minimum progress targets: [240.6 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.1962573769740719
New scales: [0.18152586 0.81847414]
5 9.77e+00 6.43e+01 4.17e+01 4.71e+02 8.76e+01 0
geophys. misfits: 73.7 (target 30.0 [False]); 21.5 (target 30.0 [True]) | smallness misfit: 1231.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [73.7 21.5]; minimum progress targets: [192.8 30. ]
Updating scaling for data misfits by 1.394424574604439
New scales: [0.23621177 0.76378823]
6 9.77e+00 3.38e+01 4.38e+01 4.62e+02 6.63e+01 0
geophys. misfits: 53.3 (target 30.0 [False]); 21.6 (target 30.0 [True]) | smallness misfit: 1197.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [53.3 21.6]; minimum progress targets: [59. 30.]
Updating scaling for data misfits by 1.3878836708388158
New scales: [0.30031847 0.69968153]
7 9.77e+00 3.11e+01 4.42e+01 4.63e+02 6.62e+01 0 Skip BFGS
geophys. misfits: 41.0 (target 30.0 [False]); 21.8 (target 30.0 [True]) | smallness misfit: 1170.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [41. 21.8]; minimum progress targets: [42.6 30. ]
Updating scaling for data misfits by 1.3765406908399755
New scales: [0.37140171 0.62859829]
8 9.77e+00 2.89e+01 4.45e+01 4.64e+02 5.68e+01 0 Skip BFGS
geophys. misfits: 33.4 (target 30.0 [False]); 22.1 (target 30.0 [True]) | smallness misfit: 1143.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [33.4 22.1]; minimum progress targets: [32.8 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.357874025735914
New scales: [0.44514965 0.55485035]
9 4.89e+00 2.71e+01 4.48e+01 2.46e+02 7.45e+01 0 Skip BFGS
geophys. misfits: 18.7 (target 30.0 [True]); 20.0 (target 30.0 [True]) | smallness misfit: 1192.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [18.7 20. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.5522062066452478
10 4.89e+00 1.94e+01 4.72e+01 2.50e+02 7.72e+01 0
geophys. misfits: 18.3 (target 30.0 [True]); 21.2 (target 30.0 [True]) | smallness misfit: 1058.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [18.3 21.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.37124802951631
11 4.89e+00 1.99e+01 4.89e+01 2.59e+02 5.77e+01 0
geophys. misfits: 17.7 (target 30.0 [True]); 22.6 (target 30.0 [True]) | smallness misfit: 939.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.7 22.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.5783204919103775
12 4.89e+00 2.04e+01 5.12e+01 2.71e+02 6.79e+01 0
geophys. misfits: 17.5 (target 30.0 [True]); 24.3 (target 30.0 [True]) | smallness misfit: 828.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.5 24.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 5.267298370992695
13 4.89e+00 2.13e+01 5.40e+01 2.85e+02 7.41e+01 0
geophys. misfits: 17.0 (target 30.0 [True]); 26.2 (target 30.0 [True]) | smallness misfit: 736.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [17. 26.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 7.652794277100461
14 4.89e+00 2.21e+01 5.76e+01 3.04e+02 9.33e+01 0
geophys. misfits: 17.3 (target 30.0 [True]); 28.9 (target 30.0 [True]) | smallness misfit: 644.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.3 28.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 10.606773691903875
15 4.89e+00 2.37e+01 6.12e+01 3.23e+02 9.25e+01 0
geophys. misfits: 17.8 (target 30.0 [True]); 32.7 (target 30.0 [False]) | smallness misfit: 554.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.8 32.7]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.6862696339324812
New scales: [0.32239071 0.67760929]
16 2.44e+00 2.79e+01 6.03e+01 1.75e+02 1.02e+02 0
geophys. misfits: 15.7 (target 30.0 [True]); 23.5 (target 30.0 [True]) | smallness misfit: 600.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [15.7 23.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 16.904836392612
17 2.44e+00 2.10e+01 7.03e+01 1.93e+02 9.47e+01 0
geophys. misfits: 16.4 (target 30.0 [True]); 25.4 (target 30.0 [True]) | smallness misfit: 486.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [16.4 25.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 25.414556384964417
18 2.44e+00 2.25e+01 7.81e+01 2.13e+02 1.02e+02 0
geophys. misfits: 21.8 (target 30.0 [True]); 22.3 (target 30.0 [True]) | smallness misfit: 419.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.8 22.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 34.573459035756194
19 2.44e+00 2.22e+01 8.42e+01 2.28e+02 1.09e+02 0
geophys. misfits: 19.6 (target 30.0 [True]); 21.4 (target 30.0 [True]) | smallness misfit: 341.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [19.6 21.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 50.69791210176808
20 2.44e+00 2.08e+01 9.46e+01 2.52e+02 1.18e+02 0
geophys. misfits: 25.7 (target 30.0 [True]); 27.4 (target 30.0 [True]) | smallness misfit: 249.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.7 27.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 57.39230776509479
21 2.44e+00 2.68e+01 9.25e+01 2.53e+02 1.08e+02 0
geophys. misfits: 33.8 (target 30.0 [False]); 31.5 (target 30.0 [False]) | smallness misfit: 243.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [33.8 31.5]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
22 1.22e+00 3.22e+01 9.02e+01 1.42e+02 1.03e+02 0
geophys. misfits: 21.8 (target 30.0 [True]); 23.7 (target 30.0 [True]) | smallness misfit: 254.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.8 23.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 75.83211639696805
23 1.22e+00 2.31e+01 1.04e+02 1.50e+02 9.58e+01 0
geophys. misfits: 24.1 (target 30.0 [True]); 23.8 (target 30.0 [True]) | smallness misfit: 206.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.1 23.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 94.83677821047651
24 1.22e+00 2.39e+01 1.10e+02 1.59e+02 1.02e+02 0
geophys. misfits: 25.5 (target 30.0 [True]); 25.2 (target 30.0 [True]) | smallness misfit: 207.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.5 25.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 112.24697235709137
25 1.22e+00 2.53e+01 1.17e+02 1.68e+02 1.07e+02 0
geophys. misfits: 25.8 (target 30.0 [True]); 26.2 (target 30.0 [True]) | smallness misfit: 187.2 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [25.8 26.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 129.68758391780855
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 4.9887e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 1.0719e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.0719e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 26
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.1
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [0.0003502883080097992, 0.0, 3.486404571820042e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09850237 0.90149763]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09850237 0.90149763]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.97e+03 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 87810.7 (target 30.0 [False]); 63893.7 (target 30.0 [False]) | smallness misfit: 282.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [87810.7 63893.7]; minimum progress targets: [240000. 240000.]
1 1.97e+03 6.62e+04 6.73e-01 6.76e+04 9.16e+01 0
geophys. misfits: 624.0 (target 30.0 [False]); 22.5 (target 30.0 [True]) | smallness misfit: 101.2 (target: 200.0 [True])
Beta cooling evaluation: progress: [624. 22.5]; minimum progress targets: [70248.6 51115. ]
Updating scaling for data misfits by 1.335112655453233
New scales: [0.12730936 0.87269064]
2 1.97e+03 9.90e+01 2.76e-01 6.45e+02 8.89e+01 0 Skip BFGS
geophys. misfits: 93.5 (target 30.0 [False]); 21.9 (target 30.0 [True]) | smallness misfit: 50.9 (target: 200.0 [True])
Beta cooling evaluation: progress: [93.5 21.9]; minimum progress targets: [499.2 30. ]
Updating scaling for data misfits by 1.3726883101291
New scales: [0.16684005 0.83315995]
3 1.97e+03 3.38e+01 1.36e-01 3.03e+02 9.63e+01 0 Skip BFGS
geophys. misfits: 64.0 (target 30.0 [False]); 21.0 (target 30.0 [True]) | smallness misfit: 47.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [64. 21.]; minimum progress targets: [74.8 30. ]
Updating scaling for data misfits by 1.4268414202187247
New scales: [0.22222847 0.77777153]
4 1.97e+03 3.06e+01 1.29e-01 2.84e+02 7.00e+01 0
geophys. misfits: 45.2 (target 30.0 [False]); 21.7 (target 30.0 [True]) | smallness misfit: 48.0 (target: 200.0 [True])
Beta cooling evaluation: progress: [45.2 21.7]; minimum progress targets: [51.2 30. ]
Updating scaling for data misfits by 1.3823203086624076
New scales: [0.28313507 0.71686493]
5 1.97e+03 2.84e+01 1.30e-01 2.85e+02 8.14e+01 0 Skip BFGS
geophys. misfits: 35.7 (target 30.0 [False]); 22.6 (target 30.0 [True]) | smallness misfit: 48.5 (target: 200.0 [True])
Beta cooling evaluation: progress: [35.7 22.6]; minimum progress targets: [36.2 30. ]
Updating scaling for data misfits by 1.3264641324828763
New scales: [0.34379075 0.65620925]
6 1.97e+03 2.71e+01 1.31e-01 2.85e+02 6.30e+01 0 Skip BFGS
geophys. misfits: 30.4 (target 30.0 [False]); 23.5 (target 30.0 [True]) | smallness misfit: 48.7 (target: 200.0 [True])
Beta cooling evaluation: progress: [30.4 23.5]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.2784008935309614
New scales: [0.40111137 0.59888863]
7 9.87e+02 2.63e+01 1.31e-01 1.56e+02 9.90e+01 0 Skip BFGS
geophys. misfits: 20.8 (target 30.0 [True]); 19.8 (target 30.0 [True]) | smallness misfit: 50.6 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [20.8 19.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.477871975281782
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 1.7688e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 9.8972e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.8972e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 8
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.1
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.136682718424762e-05, 0.0, 3.1417153861777845e-05, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09850237 0.90149763]
/home/vsts/work/1/s/simpeg/directives/directives.py:334: UserWarning:
There is no PGI regularization. Smallness target is turned off (TriggerSmall flag)
Initial data misfit scales: [0.09850237 0.90149763]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.12e+06 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 58639.3 (target 30.0 [False]); 38893.0 (target 30.0 [False])
1 2.25e+05 4.08e+04 4.00e-02 4.98e+04 1.38e+02 0
geophys. misfits: 8958.4 (target 30.0 [False]); 4393.2 (target 30.0 [False])
2 4.49e+04 4.84e+03 1.04e-01 9.53e+03 1.31e+02 0 Skip BFGS
geophys. misfits: 603.1 (target 30.0 [False]); 286.4 (target 30.0 [False])
3 8.99e+03 3.18e+02 1.40e-01 1.57e+03 1.03e+02 0 Skip BFGS
geophys. misfits: 42.3 (target 30.0 [False]); 36.9 (target 30.0 [False])
4 1.80e+03 3.74e+01 1.51e-01 3.08e+02 8.76e+01 0 Skip BFGS
geophys. misfits: 15.4 (target 30.0 [True]); 18.7 (target 30.0 [True])
All targets have been reached
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 5.4077e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 8.7562e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 8.7562e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:301: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:308: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:346: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:353: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:360: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:368: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:402: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:409: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
import discretize as Mesh
import matplotlib.pyplot as plt
import numpy as np
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 30
jk = np.linspace(1.0, 59.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
m0 = np.zeros(mesh.nC)
m0[20:41] = np.linspace(0.0, 1.0, 21)
m0[41:57] = np.linspace(-1, 0.0, 16)
poly0 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[0.0, -4.0, 4.0])
poly1 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[-0.0, 3.0, 6.0, 6.0])
poly0_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, -4.0, 4.0])
poly1_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, 3.0, 6.0, 6.0])
cluster_mapping = [maps.IdentityMap(), poly0_inverse, poly1_inverse]
m1 = np.zeros(100)
m1[20:41] = 1.0 + (poly0 * np.vstack([m0[20:41], m1[20:41]]).T)[:, 1]
m1[41:57] = -1.0 + (poly1 * np.vstack([m0[41:57], m1[41:57]]).T)[:, 1]
model2d = np.vstack([m0, m1]).T
m = utils.mkvc(model2d)
clfmapping = utils.GaussianMixtureWithNonlinearRelationships(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
means_init=np.array(
[
[0, 0],
[m0[20:41].mean(), m1[20:41].mean()],
[m0[41:57].mean(), m1[41:57].mean()],
]
),
verbose=0,
verbose_interval=10,
cluster_mapping=cluster_mapping,
)
clfmapping = clfmapping.fit(model2d)
clfnomapping = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
verbose=0,
verbose_interval=10,
)
clfnomapping = clfnomapping.fit(model2d)
wires = maps.Wires(("m1", mesh.nC), ("m2", mesh.nC))
relatrive_error = 0.01
noise_floor = 0.0
prob1 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m1)
survey1 = prob1.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
prob2 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m2)
survey2 = prob2.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
dmis1 = data_misfit.L2DataMisfit(simulation=prob1, data=survey1)
dmis2 = data_misfit.L2DataMisfit(simulation=prob2, data=survey2)
dmis = dmis1 + dmis2
minit = np.zeros_like(m)
# Distance weighting
wr1 = np.sum(prob1.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr1 = wr1 / np.max(wr1)
wr2 = np.sum(prob2.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr2 = wr2 / np.max(wr2)
reg_simple = regularization.PGI(
mesh=mesh,
gmmref=clfmapping,
gmm=clfmapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=True,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[1e6, 1e4, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_map = inv.run(minit)
# Inversion with no nonlinear mapping
reg_simple_no_map = regularization.PGI(
mesh=mesh,
gmmref=clfnomapping,
gmm=clfnomapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=False,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple_no_map, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[100.0 * np.ones(2), 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(
chiSmall=1.0, TriggerSmall=True, TriggerTheta=False, verbose=True
)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_no_map = inv.run(minit)
# WeightedLeastSquares Inversion
reg1 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m1, weights={"cell_weights": wr1}
)
reg2 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m2, weights={"cell_weights": wr2}
)
reg = reg1 + reg2
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
alpha0_ratio = np.r_[1, 1, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
beta_schedule = directives.BetaSchedule(coolingFactor=5.0, coolingRate=1)
targets = directives.MultiTargetMisfits(
TriggerSmall=False,
verbose=True,
)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, targets, beta_schedule, scaling_schedule],
)
mtik = inv.run(minit)
# Final Plot
fig, axes = plt.subplots(3, 4, figsize=(25, 15))
axes = axes.reshape(12)
left, width = 0.25, 0.5
bottom, height = 0.25, 0.5
right = left + width
top = bottom + height
axes[0].set_axis_off()
axes[0].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using true nonlinear\npetrophysical relationships"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[0].transAxes,
)
axes[1].plot(mesh.cell_centers_x, wires.m1 * mcluster_map, "b.-", ms=5, marker="v")
axes[1].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[1].set_title("Problem 1")
axes[1].legend(["Recovered Model", "True Model"], loc=1)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Property 1")
axes[2].plot(mesh.cell_centers_x, wires.m2 * mcluster_map, "r.-", ms=5, marker="v")
axes[2].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[2].set_title("Problem 2")
axes[2].legend(["Recovered Model", "True Model"], loc=1)
axes[2].set_xlabel("X")
axes[2].set_ylabel("Property 2")
x, y = np.mgrid[-1:1:0.01, -4:2:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
CS = axes[3].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[3].scatter(wires.m1 * mcluster_map, wires.m2 * mcluster_map, marker="v")
axes[3].set_title("Petrophysical Distribution")
CS.collections[0].set_label("")
axes[3].legend(["True Petrophysical Distribution", "Recovered model crossplot"])
axes[3].set_xlabel("Property 1")
axes[3].set_ylabel("Property 2")
axes[4].set_axis_off()
axes[4].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using a pure\nGaussian distribution"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[4].transAxes,
)
axes[5].plot(mesh.cell_centers_x, wires.m1 * mcluster_no_map, "b.-", ms=5, marker="v")
axes[5].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[5].set_title("Problem 1")
axes[5].legend(["Recovered Model", "True Model"], loc=1)
axes[5].set_xlabel("X")
axes[5].set_ylabel("Property 1")
axes[6].plot(mesh.cell_centers_x, wires.m2 * mcluster_no_map, "r.-", ms=5, marker="v")
axes[6].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[6].set_title("Problem 2")
axes[6].legend(["Recovered Model", "True Model"], loc=1)
axes[6].set_xlabel("X")
axes[6].set_ylabel("Property 2")
CSF = axes[7].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.5,
label="True Petro. Distribution",
)
CS = axes[7].contour(
x,
y,
np.exp(clfnomapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
500,
cmap="viridis",
linestyles="--",
label="Modeled Petro. Distribution",
)
axes[7].scatter(
wires.m1 * mcluster_no_map,
wires.m2 * mcluster_no_map,
marker="v",
label="Recovered model crossplot",
)
axes[7].set_title("Petrophysical Distribution")
axes[7].legend()
axes[7].set_xlabel("Property 1")
axes[7].set_ylabel("Property 2")
# Tikonov
axes[8].set_axis_off()
axes[8].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Least-Squares\n~Using a single cluster"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[8].transAxes,
)
axes[9].plot(mesh.cell_centers_x, wires.m1 * mtik, "b.-", ms=5, marker="v")
axes[9].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[9].set_title("Problem 1")
axes[9].legend(["Recovered Model", "True Model"], loc=1)
axes[9].set_xlabel("X")
axes[9].set_ylabel("Property 1")
axes[10].plot(mesh.cell_centers_x, wires.m2 * mtik, "r.-", ms=5, marker="v")
axes[10].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[10].set_title("Problem 2")
axes[10].legend(["Recovered Model", "True Model"], loc=1)
axes[10].set_xlabel("X")
axes[10].set_ylabel("Property 2")
CS = axes[11].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[11].scatter(wires.m1 * mtik, wires.m2 * mtik, marker="v")
axes[11].set_title("Petro Distribution")
CS.collections[0].set_label("")
axes[11].legend(["True Petro Distribution", "Recovered model crossplot"])
axes[11].set_xlabel("Property 1")
axes[11].set_ylabel("Property 2")
plt.subplots_adjust(wspace=0.3, hspace=0.3, top=0.85)
plt.show()
Total running time of the script: (0 minutes 34.766 seconds)
Estimated memory usage: 9 MB