Note
Go to the end to download the full example code.
Magnetic Amplitude inversion on a TreeMesh#
In this example, we demonstrate the use of magnetic amplitude inversion on 3D TreeMesh for the inversion of Total Magnetic Intensity (TMI) data affected by remanence. The original idea must be credited to Shearer and Li (2005) @ CSM
First we invert the TMI for an equivalent source layer, from which we recover 3-component magnetic data. This data is then transformed to amplitude
Secondly, we invert the non-linear inverse problem with
simpeg.directives.UpdateSensitivityWeights. We also
uses the simpeg.regularization.Sparse to apply sparsity
assumption in order to improve the recovery of a compact prism.
import scipy as sp
import numpy as np
import matplotlib.pyplot as plt
from simpeg import (
data,
data_misfit,
directives,
maps,
inverse_problem,
optimization,
inversion,
regularization,
)
from simpeg.potential_fields import magnetics
from simpeg import utils
from simpeg.utils import mkvc
from discretize.utils import mesh_builder_xyz, refine_tree_xyz, active_from_xyz
# sphinx_gallery_thumbnail_number = 4
Setup#
Define the survey and model parameters
First we need to define the direction of the inducing field As a simple case, we pick a vertical inducing field of magnitude 50,000 nT.
# We will assume a vertical inducing field
h0_amplitude, h0_inclination, h0_declination = (50000.0, 90.0, 0.0)
# The magnetization is set along a different direction (induced + remanence)
M = np.array([45.0, 90.0])
# Block with an effective susceptibility
chi_e = 0.05
# Create grid of points for topography
# Lets create a simple Gaussian topo and set the active cells
[xx, yy] = np.meshgrid(np.linspace(-200, 200, 50), np.linspace(-200, 200, 50))
b = 100
A = 50
zz = A * np.exp(-0.5 * ((xx / b) ** 2.0 + (yy / b) ** 2.0))
topo = np.c_[mkvc(xx), mkvc(yy), mkvc(zz)]
# Create and array of observation points
xr = np.linspace(-100.0, 100.0, 20)
yr = np.linspace(-100.0, 100.0, 20)
X, Y = np.meshgrid(xr, yr)
Z = A * np.exp(-0.5 * ((X / b) ** 2.0 + (Y / b) ** 2.0)) + 10
# Create a MAGsurvey
rxLoc = np.c_[mkvc(X.T), mkvc(Y.T), mkvc(Z.T)]
receiver_list = magnetics.receivers.Point(rxLoc)
srcField = magnetics.sources.UniformBackgroundField(
receiver_list=[receiver_list],
amplitude=h0_amplitude,
inclination=h0_inclination,
declination=h0_declination,
)
survey = magnetics.survey.Survey(srcField)
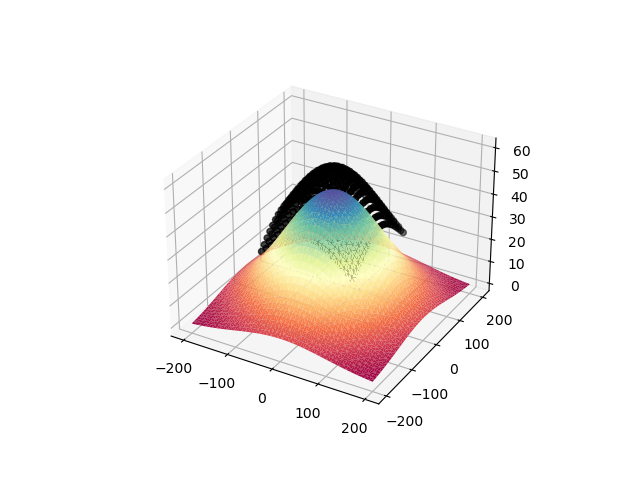
# Here how the topography looks with a quick interpolation, just a Gaussian...
tri = sp.spatial.Delaunay(topo)
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1, projection="3d")
ax.plot_trisurf(
topo[:, 0], topo[:, 1], topo[:, 2], triangles=tri.simplices, cmap=plt.cm.Spectral
)
ax.scatter3D(rxLoc[:, 0], rxLoc[:, 1], rxLoc[:, 2], c="k")
plt.show()
Inversion Mesh#
Here, we create a TreeMesh with base cell size of 5 m. We created a small utility function to center the mesh around points and to figure out the outermost dimension for adequate padding distance. The second stage allows us to refine the mesh around points or surfaces (point assumed to follow an horiontal interface such as topo)
# Create a mesh
h = [5, 5, 5]
padDist = np.ones((3, 2)) * 100
mesh = mesh_builder_xyz(
rxLoc, h, padding_distance=padDist, depth_core=100, mesh_type="tree"
)
mesh = refine_tree_xyz(
mesh, topo, method="surface", octree_levels=[4, 4], finalize=True
)
# Define the active cells from topo
actv = active_from_xyz(mesh, topo)
nC = int(actv.sum())
/home/vsts/work/1/s/examples/03-magnetics/plot_inv_mag_nonLinear_Amplitude.py:114: DeprecationWarning:
The surface option is deprecated as of `0.9.0` please update your code to use the `TreeMesh.refine_surface` functionality. It will be removed in a future version of discretize.
Forward modeling data#
We can now generate TMI data
# Convert the inclination and declination to vector in Cartesian
M_xyz = utils.mat_utils.dip_azimuth2cartesian(np.ones(nC) * M[0], np.ones(nC) * M[1])
# Get the indicies of the magnetized block
ind = utils.model_builder.get_indices_block(
np.r_[-20, -20, -10],
np.r_[20, 20, 25],
mesh.gridCC,
)[0]
# Assign magnetization value, inducing field strength will
# be applied in by the :class:`simpeg.PF.Magnetics` problem
model = np.zeros(mesh.nC)
model[ind] = chi_e
# Remove air cells
model = model[actv]
# Create reduced identity map
idenMap = maps.IdentityMap(nP=nC)
# Create the forward model operator
simulation = magnetics.simulation.Simulation3DIntegral(
survey=survey,
mesh=mesh,
chiMap=idenMap,
ind_active=actv,
store_sensitivities="forward_only",
)
simulation.M = M_xyz
# Compute some data and add some random noise
synthetic_data = simulation.dpred(model)
# Split the data in components
nD = rxLoc.shape[0]
std = 5 # nT
synthetic_data += np.random.randn(nD) * std
wd = np.ones(nD) * std
# Assign data and uncertainties to the survey
data_object = data.Data(survey, dobs=synthetic_data, standard_deviation=wd)
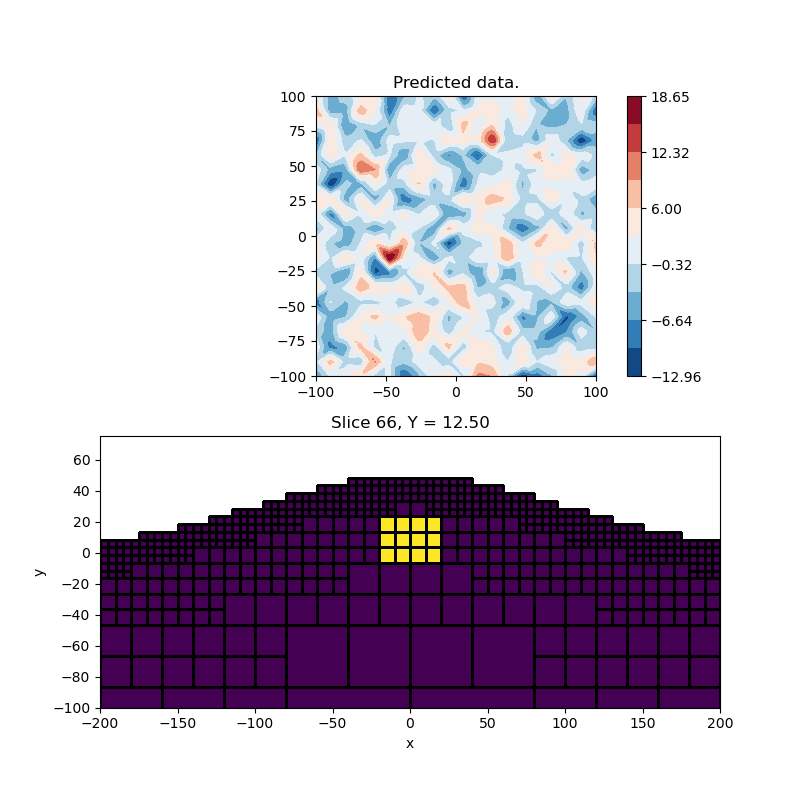
# Plot the model and data
plt.figure(figsize=(8, 8))
ax = plt.subplot(2, 1, 1)
im = utils.plot_utils.plot2Ddata(
rxLoc, synthetic_data, ax=ax, contourOpts={"cmap": "RdBu_r"}
)
plt.colorbar(im[0])
ax.set_title("Predicted data.")
plt.gca().set_aspect("equal", adjustable="box")
# Plot the vector model
ax = plt.subplot(2, 1, 2)
# Create active map to go from reduce set to full
actvPlot = maps.InjectActiveCells(mesh, actv, np.nan)
mesh.plot_slice(
actvPlot * model,
ax=ax,
normal="Y",
ind=66,
pcolor_opts={"vmin": 0.0, "vmax": 0.01},
grid=True,
)
ax.set_xlim([-200, 200])
ax.set_ylim([-100, 75])
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.gca().set_aspect("equal", adjustable="box")
plt.show()
Equivalent Source#
We first need to convert the TMI data into amplitude. We do this by an effective susceptibility layer, from which we can forward component data
# Get the active cells for equivalent source is the topo only
surf = active_from_xyz(mesh, topo)
nC = np.count_nonzero(surf) # Number of active cells
mstart = np.ones(nC) * 1e-4
# Create active map to go from reduce set to full
surfMap = maps.InjectActiveCells(mesh, surf, np.nan)
# Create identity map
idenMap = maps.IdentityMap(nP=nC)
# Create static map
simulation = magnetics.simulation.Simulation3DIntegral(
mesh=mesh, survey=survey, chiMap=idenMap, ind_active=surf, store_sensitivities="ram"
)
wr = simulation.getJtJdiag(mstart) ** 0.5
wr = wr / np.max(np.abs(wr))
# Create a regularization function, in this case l2l2
reg = regularization.Sparse(
mesh, active_cells=surf, mapping=maps.IdentityMap(nP=nC), alpha_z=0
)
reg.reference_model = np.zeros(nC)
# Specify how the optimization will proceed, set susceptibility bounds to inf
opt = optimization.ProjectedGNCG(
maxIter=20, lower=-np.inf, upper=np.inf, maxIterLS=20, maxIterCG=20, tolCG=1e-3
)
# Define misfit function (obs-calc)
dmis = data_misfit.L2DataMisfit(simulation=simulation, data=data_object)
# Create the default L2 inverse problem from the above objects
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# Specify how the initial beta is found
betaest = directives.BetaEstimate_ByEig(beta0_ratio=2)
# Target misfit to stop the inversion,
# try to fit as much as possible of the signal, we don't want to lose anything
IRLS = directives.Update_IRLS(
f_min_change=1e-3, minGNiter=1, beta_tol=1e-1, max_irls_iterations=5
)
update_Jacobi = directives.UpdatePreconditioner()
# Put all the parts together
inv = inversion.BaseInversion(invProb, directiveList=[betaest, IRLS, update_Jacobi])
# Run the equivalent source inversion
mrec = inv.run(mstart)
Running inversion with SimPEG v0.22.2
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 7.67e+01 4.85e+02 1.36e+00 5.89e+02 1.52e+05 0
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 0.0068745558246882015
1 3.83e+01 9.35e+01 1.48e+00 1.50e+02 5.06e+02 0
2 1.32e+02 8.22e+01 2.38e+00 3.95e+02 8.55e+03 0 Skip BFGS
3 2.72e+02 1.88e+02 4.70e-01 3.16e+02 4.07e+03 0
4 5.03e+02 2.35e+02 1.90e-01 3.31e+02 3.30e+03 0 Skip BFGS
5 8.79e+02 2.67e+02 9.39e-02 3.50e+02 3.54e+03 0 Skip BFGS
Reach maximum number of IRLS cycles: 5
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 5.9014e+01
1 : |xc-x_last| = 4.4844e-03 <= tolX*(1+|x0|) = 1.0195e-01
0 : |proj(x-g)-x| = 3.5435e+03 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 3.5435e+03 <= 1e3*eps = 1.0000e-02
0 : maxIter = 20 <= iter = 6
------------------------- DONE! -------------------------
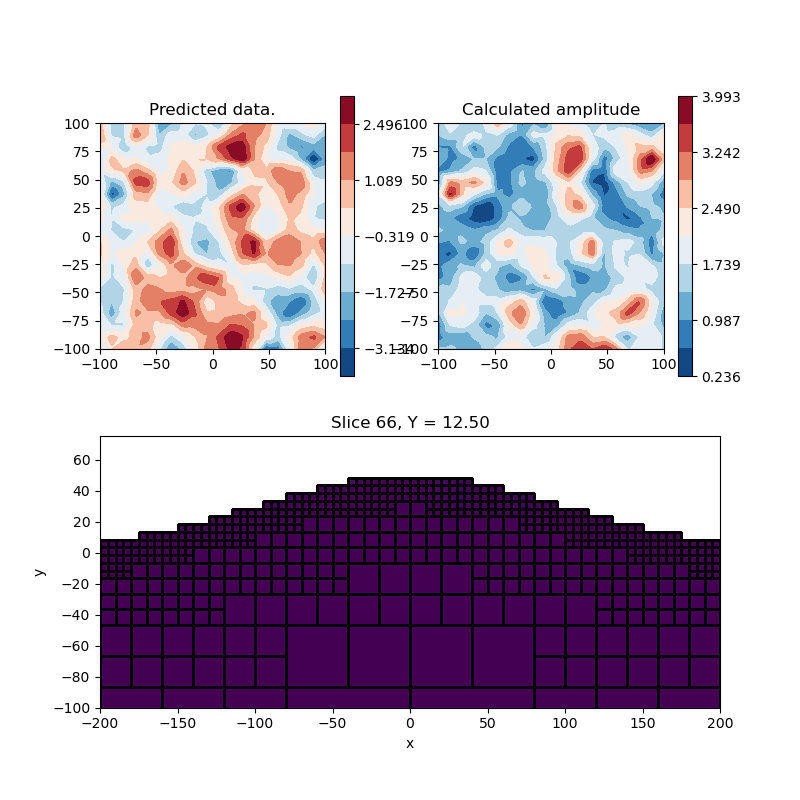
Forward Amplitude Data#
Now that we have an equialent source layer, we can forward model all three components of the field and add them up: \(|B| = \sqrt{( Bx^2 + Bx^2 + Bx^2 )}\)
receiver_list = magnetics.receivers.Point(rxLoc, components=["bx", "by", "bz"])
srcField = magnetics.sources.UniformBackgroundField(
receiver_list=[receiver_list],
amplitude=h0_amplitude,
inclination=h0_inclination,
declination=h0_declination,
)
surveyAmp = magnetics.survey.Survey(srcField)
simulation = magnetics.simulation.Simulation3DIntegral(
mesh=mesh, survey=surveyAmp, chiMap=idenMap, ind_active=surf, is_amplitude_data=True
)
bAmp = simulation.fields(mrec)
# Plot the layer model and data
plt.figure(figsize=(8, 8))
ax = plt.subplot(2, 2, 1)
im = utils.plot_utils.plot2Ddata(
rxLoc, invProb.dpred, ax=ax, contourOpts={"cmap": "RdBu_r"}
)
plt.colorbar(im[0])
ax.set_title("Predicted data.")
plt.gca().set_aspect("equal", adjustable="box")
ax = plt.subplot(2, 2, 2)
im = utils.plot_utils.plot2Ddata(rxLoc, bAmp, ax=ax, contourOpts={"cmap": "RdBu_r"})
plt.colorbar(im[0])
ax.set_title("Calculated amplitude")
plt.gca().set_aspect("equal", adjustable="box")
# Plot the equivalent layer model
ax = plt.subplot(2, 1, 2)
mesh.plot_slice(
surfMap * mrec,
ax=ax,
normal="Y",
ind=66,
pcolor_opts={"vmin": 0.0, "vmax": 0.01},
grid=True,
)
ax.set_xlim([-200, 200])
ax.set_ylim([-100, 75])
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.gca().set_aspect("equal", adjustable="box")
plt.show()
Amplitude Inversion#
Now that we have amplitude data, we can invert for an effective susceptibility. This is a non-linear inversion.
# Create active map to go from reduce space to full
actvMap = maps.InjectActiveCells(mesh, actv, -100)
nC = int(actv.sum())
# Create identity map
idenMap = maps.IdentityMap(nP=nC)
mstart = np.ones(nC) * 1e-4
# Create the forward model operator
simulation = magnetics.simulation.Simulation3DIntegral(
survey=surveyAmp, mesh=mesh, chiMap=idenMap, ind_active=actv, is_amplitude_data=True
)
data_obj = data.Data(survey, dobs=bAmp, noise_floor=wd)
# Create a sparse regularization
reg = regularization.Sparse(mesh, active_cells=actv, mapping=idenMap)
reg.norms = [1, 0, 0, 0]
reg.reference_model = np.zeros(nC)
# Data misfit function
dmis = data_misfit.L2DataMisfit(simulation=simulation, data=data_obj)
# Add directives to the inversion
opt = optimization.ProjectedGNCG(
maxIter=30, lower=0.0, upper=1.0, maxIterLS=20, maxIterCG=20, tolCG=1e-3
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# Here is the list of directives
betaest = directives.BetaEstimate_ByEig(beta0_ratio=1)
# Specify the sparse norms
IRLS = directives.Update_IRLS(
max_irls_iterations=10,
f_min_change=1e-3,
minGNiter=1,
coolingRate=1,
beta_search=False,
)
# Special directive specific to the mag amplitude problem. The sensitivity
# weights are updated between each iteration.
update_SensWeight = directives.UpdateSensitivityWeights()
update_Jacobi = directives.UpdatePreconditioner()
# Put all together
inv = inversion.BaseInversion(
invProb, directiveList=[update_SensWeight, betaest, IRLS, update_Jacobi]
)
# Invert
mrec_Amp = inv.run(mstart)
Running inversion with SimPEG v0.22.2
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 2.03e+04 1.43e+01 3.71e-03 8.98e+01 6.98e+01 0
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 7.62418358472046e-05
1 1.02e+04 9.57e+00 3.02e-04 1.26e+01 9.93e+01 0
2 1.18e+06 5.22e+00 6.82e-04 8.10e+02 5.43e+01 0
3 1.62e+07 4.71e+01 1.42e-06 7.01e+01 4.09e+01 0
4 2.06e+08 5.12e+01 1.55e-08 5.44e+01 3.23e+01 0 Skip BFGS
5 2.60e+09 5.17e+01 1.07e-10 5.20e+01 2.55e+01 0
6 3.28e+10 5.17e+01 6.99e-13 5.18e+01 3.14e+01 0
7 4.13e+11 5.17e+01 4.57e-15 5.17e+01 3.84e+01 0
8 5.20e+12 5.17e+01 2.99e-17 5.17e+01 2.63e+01 0
9 6.55e+13 5.17e+01 1.89e-19 5.17e+01 1.40e-10 0
------------------------- STOP! -------------------------
1 : |fc-fOld| = 1.2034e-04 <= tolF*(1+|f0|) = 9.0768e+00
1 : |xc-x_last| = 1.6164e-09 <= tolX*(1+|x0|) = 1.0195e-01
1 : |proj(x-g)-x| = 1.3951e-10 <= tolG = 1.0000e-01
1 : |proj(x-g)-x| = 1.3951e-10 <= 1e3*eps = 1.0000e-02
0 : maxIter = 30 <= iter = 9
------------------------- DONE! -------------------------
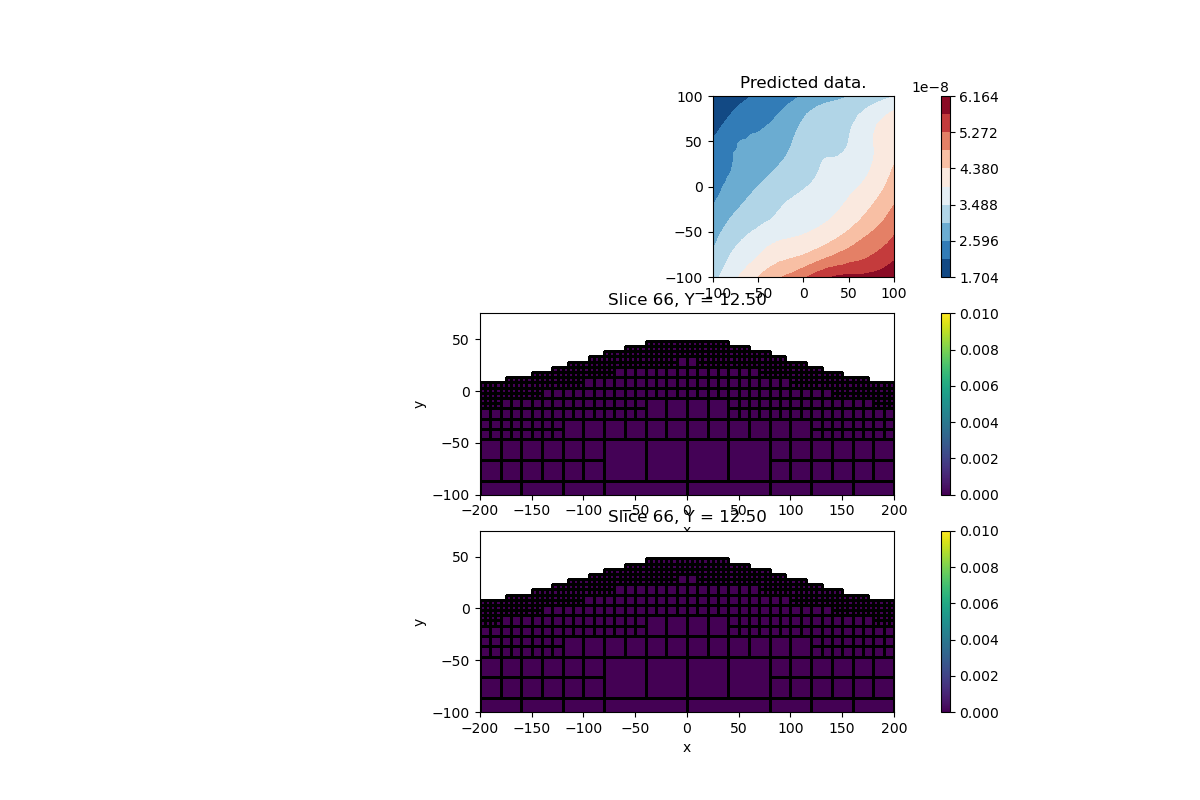
Final Plot#
Let’s compare the smooth and compact model Note that the recovered effective susceptibility block is slightly offset to the left of the true model. This is due to the wrong assumption of a vertical magnetization. Important to remember that the amplitude inversion is weakly sensitive to the magnetization direction, but can still have an impact.
# Plot the layer model and data
plt.figure(figsize=(12, 8))
ax = plt.subplot(3, 1, 1)
im = utils.plot_utils.plot2Ddata(
rxLoc, invProb.dpred, ax=ax, contourOpts={"cmap": "RdBu_r"}
)
plt.colorbar(im[0])
ax.set_title("Predicted data.")
plt.gca().set_aspect("equal", adjustable="box")
# Plot the l2 model
ax = plt.subplot(3, 1, 2)
im = mesh.plot_slice(
actvPlot * invProb.l2model,
ax=ax,
normal="Y",
ind=66,
pcolor_opts={"vmin": 0.0, "vmax": 0.01},
grid=True,
)
plt.colorbar(im[0])
ax.set_xlim([-200, 200])
ax.set_ylim([-100, 75])
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.gca().set_aspect("equal", adjustable="box")
# Plot the lp model
ax = plt.subplot(3, 1, 3)
im = mesh.plot_slice(
actvPlot * invProb.model,
ax=ax,
normal="Y",
ind=66,
pcolor_opts={"vmin": 0.0, "vmax": 0.01},
grid=True,
)
plt.colorbar(im[0])
ax.set_xlim([-200, 200])
ax.set_ylim([-100, 75])
ax.set_xlabel("x")
ax.set_ylabel("y")
plt.gca().set_aspect("equal", adjustable="box")
plt.show()
Total running time of the script: (0 minutes 36.938 seconds)
Estimated memory usage: 688 MB