Note
Go to the end to download the full example code.
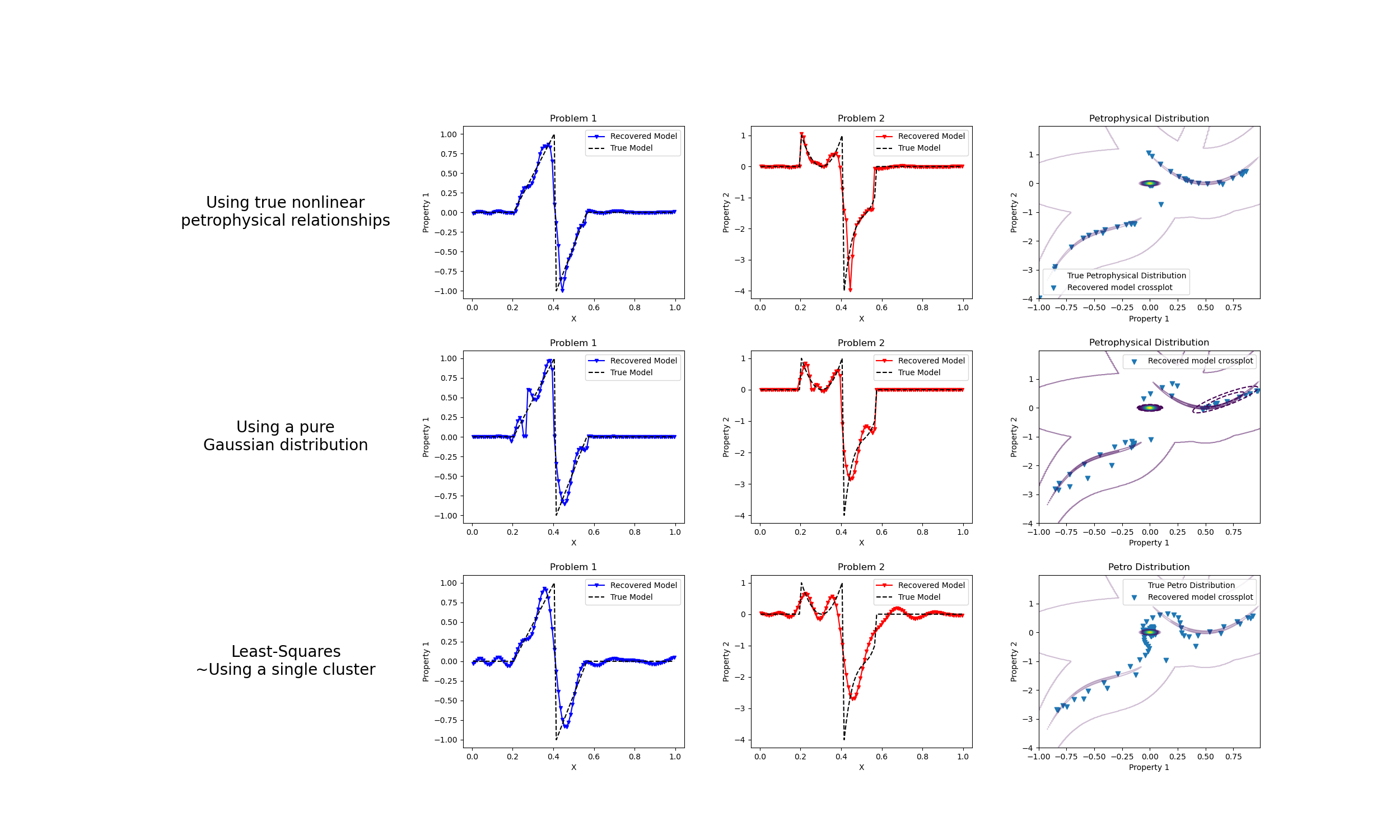
Petrophysically guided inversion: Joint linear example with nonlinear relationships#
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically guided inversion. We explore it through coupling two linear problems whose respective physical properties are linked by polynomial relationships that change between rock units.
Running inversion with SimPEG v0.22.2
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [4.700617082979148, 0.0, 3.7307624261940927e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.10047705 0.89952295]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.10047705 0.89952295]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.46e+01 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 887.8 (target 30.0 [False]); 57.0 (target 30.0 [False]) | smallness misfit: 3029.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [887.8 57. ]; minimum progress targets: [240000. 240000.]
1 1.46e+01 1.41e+02 5.40e+01 9.27e+02 7.77e+01 0
geophys. misfits: 484.4 (target 30.0 [False]); 22.3 (target 30.0 [True]) | smallness misfit: 1493.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [484.4 22.3]; minimum progress targets: [710.2 45.6]
Updating scaling for data misfits by 1.3459613509227142
New scales: [0.13069511 0.86930489]
2 1.46e+01 8.27e+01 5.30e+01 8.55e+02 9.57e+01 0 Skip BFGS
geophys. misfits: 327.9 (target 30.0 [False]); 22.4 (target 30.0 [True]) | smallness misfit: 1402.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [327.9 22.4]; minimum progress targets: [387.5 30. ]
Updating scaling for data misfits by 1.3400703841491697
New scales: [0.16768768 0.83231232]
3 1.46e+01 7.36e+01 5.42e+01 8.63e+02 6.86e+01 0 Skip BFGS
geophys. misfits: 227.6 (target 30.0 [False]); 22.3 (target 30.0 [True]) | smallness misfit: 1346.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [227.6 22.3]; minimum progress targets: [262.3 30. ]
Updating scaling for data misfits by 1.346549268195412
New scales: [0.21339869 0.78660131]
4 1.46e+01 6.61e+01 5.52e+01 8.70e+02 7.47e+01 0 Skip BFGS
geophys. misfits: 161.0 (target 30.0 [False]); 22.5 (target 30.0 [True]) | smallness misfit: 1297.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [161. 22.5]; minimum progress targets: [182.1 30. ]
Updating scaling for data misfits by 1.3351472444242103
New scales: [0.26590141 0.73409859]
5 1.46e+01 5.93e+01 5.61e+01 8.76e+02 7.22e+01 0
geophys. misfits: 118.4 (target 30.0 [False]); 22.6 (target 30.0 [True]) | smallness misfit: 1262.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [118.4 22.6]; minimum progress targets: [128.8 30. ]
Updating scaling for data misfits by 1.3298669091383504
New scales: [0.32509843 0.67490157]
6 1.46e+01 5.37e+01 5.67e+01 8.80e+02 6.98e+01 0 Skip BFGS
geophys. misfits: 90.5 (target 30.0 [False]); 22.8 (target 30.0 [True]) | smallness misfit: 1232.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [90.5 22.8]; minimum progress targets: [94.7 30. ]
Updating scaling for data misfits by 1.3177391848296285
New scales: [0.38828632 0.61171368]
7 1.46e+01 4.91e+01 5.73e+01 8.84e+02 8.43e+01 0 Skip BFGS
geophys. misfits: 72.0 (target 30.0 [False]); 23.2 (target 30.0 [True]) | smallness misfit: 1203.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [72. 23.2]; minimum progress targets: [72.4 30. ]
Updating scaling for data misfits by 1.2941255077113822
New scales: [0.45098637 0.54901363]
8 1.46e+01 4.52e+01 5.77e+01 8.86e+02 7.57e+01 0 Skip BFGS
geophys. misfits: 60.0 (target 30.0 [False]); 23.7 (target 30.0 [True]) | smallness misfit: 1182.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [60. 23.7]; minimum progress targets: [57.6 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.2657478031432277
New scales: [0.509743 0.490257]
9 7.28e+00 4.22e+01 5.80e+01 4.65e+02 9.73e+01 0 Skip BFGS
geophys. misfits: 27.7 (target 30.0 [True]); 20.4 (target 30.0 [True]) | smallness misfit: 1205.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [27.7 20.4]; minimum progress targets: [48. 30.]
Warming alpha_pgi to favor clustering: 1.2752371504641618
10 7.28e+00 2.42e+01 6.03e+01 4.64e+02 7.38e+01 0
geophys. misfits: 27.5 (target 30.0 [True]); 21.2 (target 30.0 [True]) | smallness misfit: 1142.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [27.5 21.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.5978844764258795
11 7.28e+00 2.44e+01 6.11e+01 4.69e+02 5.91e+01 0
geophys. misfits: 27.1 (target 30.0 [True]); 22.2 (target 30.0 [True]) | smallness misfit: 1079.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [27.1 22.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.9653309153742942
12 7.28e+00 2.47e+01 6.19e+01 4.75e+02 4.28e+01 0
geophys. misfits: 26.7 (target 30.0 [True]); 23.2 (target 30.0 [True]) | smallness misfit: 1019.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.7 23.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.3715113109250527
13 7.28e+00 2.50e+01 6.27e+01 4.82e+02 5.03e+01 0
geophys. misfits: 26.5 (target 30.0 [True]); 24.4 (target 30.0 [True]) | smallness misfit: 965.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.5 24.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.803894374640154
14 7.28e+00 2.54e+01 6.36e+01 4.88e+02 7.89e+01 0
geophys. misfits: 26.1 (target 30.0 [True]); 25.7 (target 30.0 [True]) | smallness misfit: 912.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.1 25.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.24861450620946
15 7.28e+00 2.59e+01 6.44e+01 4.95e+02 8.11e+01 0
geophys. misfits: 25.9 (target 30.0 [True]); 27.0 (target 30.0 [True]) | smallness misfit: 868.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.9 27. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.6835682108627106
16 7.28e+00 2.65e+01 6.51e+01 5.01e+02 7.80e+01 0
geophys. misfits: 25.9 (target 30.0 [True]); 28.3 (target 30.0 [True]) | smallness misfit: 826.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.9 28.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 4.0869138002317165
17 7.28e+00 2.71e+01 6.57e+01 5.06e+02 8.47e+01 0
geophys. misfits: 25.7 (target 30.0 [True]); 29.7 (target 30.0 [True]) | smallness misfit: 788.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.7 29.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 4.451036376529652
18 7.28e+00 2.77e+01 6.63e+01 5.10e+02 7.63e+01 0 Skip BFGS
geophys. misfits: 25.6 (target 30.0 [True]); 30.9 (target 30.0 [False]) | smallness misfit: 759.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.6 30.9]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.1707116022801967
New scales: [0.47037603 0.52962397]
19 3.64e+00 2.84e+01 6.62e+01 2.69e+02 9.43e+01 0 Skip BFGS
geophys. misfits: 18.9 (target 30.0 [True]); 23.1 (target 30.0 [True]) | smallness misfit: 829.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [18.9 23.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 6.4349061606984135
20 3.64e+00 2.11e+01 7.11e+01 2.80e+02 8.13e+01 0
geophys. misfits: 18.8 (target 30.0 [True]); 25.3 (target 30.0 [True]) | smallness misfit: 714.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [18.8 25.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 8.93690027510985
21 3.64e+00 2.23e+01 7.45e+01 2.94e+02 8.39e+01 0
geophys. misfits: 17.6 (target 30.0 [True]); 24.0 (target 30.0 [True]) | smallness misfit: 658.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.6 24. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 13.213503393977751
22 3.64e+00 2.10e+01 8.09e+01 3.15e+02 9.81e+01 0
geophys. misfits: 16.9 (target 30.0 [True]); 23.8 (target 30.0 [True]) | smallness misfit: 575.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [16.9 23.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 20.091711694672696
23 3.64e+00 2.05e+01 8.93e+01 3.46e+02 1.07e+02 0
geophys. misfits: 17.6 (target 30.0 [True]); 24.4 (target 30.0 [True]) | smallness misfit: 509.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.6 24.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 29.466249029788962
24 3.64e+00 2.12e+01 9.83e+01 3.79e+02 1.18e+02 0
geophys. misfits: 17.8 (target 30.0 [True]); 25.0 (target 30.0 [True]) | smallness misfit: 399.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [17.8 25. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 42.46995844548629
25 3.64e+00 2.16e+01 1.08e+02 4.13e+02 1.17e+02 0
geophys. misfits: 21.7 (target 30.0 [True]); 27.7 (target 30.0 [True]) | smallness misfit: 322.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.7 27.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 52.28185160467312
26 3.64e+00 2.49e+01 1.11e+02 4.29e+02 1.28e+02 0
geophys. misfits: 26.7 (target 30.0 [True]); 38.6 (target 30.0 [False]) | smallness misfit: 254.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.7 38.6]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.1220001990653226
New scales: [0.44182768 0.55817232]
27 1.82e+00 3.33e+01 1.06e+02 2.26e+02 1.10e+02 0
geophys. misfits: 19.6 (target 30.0 [True]); 28.6 (target 30.0 [True]) | smallness misfit: 273.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [19.6 28.6]; minimum progress targets: [30. 30.9]
Warming alpha_pgi to favor clustering: 67.39823687526464
28 1.82e+00 2.46e+01 1.17e+02 2.38e+02 1.05e+02 0
geophys. misfits: 19.3 (target 30.0 [True]); 27.6 (target 30.0 [True]) | smallness misfit: 255.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [19.3 27.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 88.93270867488775
29 1.82e+00 2.39e+01 1.29e+02 2.59e+02 1.20e+02 0
geophys. misfits: 20.8 (target 30.0 [True]); 29.0 (target 30.0 [True]) | smallness misfit: 208.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [20.8 29. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 110.09965074361558
30 1.82e+00 2.54e+01 1.35e+02 2.71e+02 1.16e+02 0
geophys. misfits: 21.9 (target 30.0 [True]); 28.6 (target 30.0 [True]) | smallness misfit: 183.6 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [21.9 28.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 133.05122914742725
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 3.3437e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 1.1599e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.1599e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 31
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.2
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [0.00034585937269245243, 0.0, 3.463789898428949e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.10047705 0.89952295]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.10047705 0.89952295]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.98e+03 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 86876.3 (target 30.0 [False]); 63978.4 (target 30.0 [False]) | smallness misfit: 294.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [86876.3 63978.4]; minimum progress targets: [240000. 240000.]
1 1.98e+03 6.63e+04 6.89e-01 6.76e+04 9.06e+01 0
geophys. misfits: 599.1 (target 30.0 [False]); 22.6 (target 30.0 [True]) | smallness misfit: 96.0 (target: 200.0 [True])
Beta cooling evaluation: progress: [599.1 22.6]; minimum progress targets: [69501. 51182.7]
Updating scaling for data misfits by 1.3270556908502626
New scales: [0.12909633 0.87090367]
2 1.98e+03 9.70e+01 2.51e-01 5.93e+02 8.69e+01 0 Skip BFGS
geophys. misfits: 86.7 (target 30.0 [False]); 21.9 (target 30.0 [True]) | smallness misfit: 47.8 (target: 200.0 [True])
Beta cooling evaluation: progress: [86.7 21.9]; minimum progress targets: [479.3 30. ]
Updating scaling for data misfits by 1.3704729990116888
New scales: [0.16884762 0.83115238]
3 1.98e+03 3.28e+01 1.34e-01 2.97e+02 8.78e+01 0 Skip BFGS
geophys. misfits: 58.6 (target 30.0 [False]); 20.8 (target 30.0 [True]) | smallness misfit: 46.4 (target: 200.0 [True])
Beta cooling evaluation: progress: [58.6 20.8]; minimum progress targets: [69.3 30. ]
Updating scaling for data misfits by 1.4405648008978333
New scales: [0.2263948 0.7736052]
4 1.98e+03 2.94e+01 1.27e-01 2.80e+02 7.14e+01 0
geophys. misfits: 41.9 (target 30.0 [False]); 21.6 (target 30.0 [True]) | smallness misfit: 47.0 (target: 200.0 [True])
Beta cooling evaluation: progress: [41.9 21.6]; minimum progress targets: [46.9 30. ]
Updating scaling for data misfits by 1.3906641446422472
New scales: [0.28925607 0.71074393]
5 1.98e+03 2.74e+01 1.28e-01 2.81e+02 6.84e+01 0 Skip BFGS
geophys. misfits: 33.0 (target 30.0 [False]); 22.3 (target 30.0 [True]) | smallness misfit: 47.4 (target: 200.0 [True])
Beta cooling evaluation: progress: [33. 22.3]; minimum progress targets: [33.5 30. ]
Updating scaling for data misfits by 1.3450563296196518
New scales: [0.35375732 0.64624268]
6 1.98e+03 2.61e+01 1.29e-01 2.81e+02 6.45e+01 0 Skip BFGS
geophys. misfits: 28.1 (target 30.0 [True]); 23.2 (target 30.0 [True]) | smallness misfit: 47.6 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [28.1 23.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.1815223852701826
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 3.3749e-02 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 6.4497e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 6.4497e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 7
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.2
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.489065259898251e-05, 0.0, 3.497730681430686e-05, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.10047705 0.89952295]
/home/vsts/work/1/s/simpeg/directives/directives.py:334: UserWarning:
There is no PGI regularization. Smallness target is turned off (TriggerSmall flag)
Initial data misfit scales: [0.10047705 0.89952295]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.06e+06 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 55098.4 (target 30.0 [False]); 36785.5 (target 30.0 [False])
1 2.12e+05 3.86e+04 4.20e-02 4.75e+04 1.38e+02 0
geophys. misfits: 8018.9 (target 30.0 [False]); 4037.1 (target 30.0 [False])
2 4.25e+04 4.44e+03 1.06e-01 8.94e+03 1.30e+02 0 Skip BFGS
geophys. misfits: 529.1 (target 30.0 [False]); 266.3 (target 30.0 [False])
3 8.50e+03 2.93e+02 1.41e-01 1.49e+03 1.03e+02 0 Skip BFGS
geophys. misfits: 41.2 (target 30.0 [False]); 34.4 (target 30.0 [False])
4 1.70e+03 3.51e+01 1.51e-01 2.92e+02 9.27e+01 0 Skip BFGS
geophys. misfits: 18.2 (target 30.0 [True]); 16.8 (target 30.0 [True])
All targets have been reached
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 4.7350e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 9.2644e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.2644e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:301: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:308: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:346: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:353: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:360: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:368: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:402: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:409: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
import discretize as Mesh
import matplotlib.pyplot as plt
import numpy as np
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 30
jk = np.linspace(1.0, 59.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
m0 = np.zeros(mesh.nC)
m0[20:41] = np.linspace(0.0, 1.0, 21)
m0[41:57] = np.linspace(-1, 0.0, 16)
poly0 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[0.0, -4.0, 4.0])
poly1 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[-0.0, 3.0, 6.0, 6.0])
poly0_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, -4.0, 4.0])
poly1_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, 3.0, 6.0, 6.0])
cluster_mapping = [maps.IdentityMap(), poly0_inverse, poly1_inverse]
m1 = np.zeros(100)
m1[20:41] = 1.0 + (poly0 * np.vstack([m0[20:41], m1[20:41]]).T)[:, 1]
m1[41:57] = -1.0 + (poly1 * np.vstack([m0[41:57], m1[41:57]]).T)[:, 1]
model2d = np.vstack([m0, m1]).T
m = utils.mkvc(model2d)
clfmapping = utils.GaussianMixtureWithNonlinearRelationships(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
means_init=np.array(
[
[0, 0],
[m0[20:41].mean(), m1[20:41].mean()],
[m0[41:57].mean(), m1[41:57].mean()],
]
),
verbose=0,
verbose_interval=10,
cluster_mapping=cluster_mapping,
)
clfmapping = clfmapping.fit(model2d)
clfnomapping = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
verbose=0,
verbose_interval=10,
)
clfnomapping = clfnomapping.fit(model2d)
wires = maps.Wires(("m1", mesh.nC), ("m2", mesh.nC))
relatrive_error = 0.01
noise_floor = 0.0
prob1 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m1)
survey1 = prob1.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
prob2 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m2)
survey2 = prob2.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
dmis1 = data_misfit.L2DataMisfit(simulation=prob1, data=survey1)
dmis2 = data_misfit.L2DataMisfit(simulation=prob2, data=survey2)
dmis = dmis1 + dmis2
minit = np.zeros_like(m)
# Distance weighting
wr1 = np.sum(prob1.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr1 = wr1 / np.max(wr1)
wr2 = np.sum(prob2.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr2 = wr2 / np.max(wr2)
reg_simple = regularization.PGI(
mesh=mesh,
gmmref=clfmapping,
gmm=clfmapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=True,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[1e6, 1e4, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_map = inv.run(minit)
# Inversion with no nonlinear mapping
reg_simple_no_map = regularization.PGI(
mesh=mesh,
gmmref=clfnomapping,
gmm=clfnomapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=False,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple_no_map, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[100.0 * np.ones(2), 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(
chiSmall=1.0, TriggerSmall=True, TriggerTheta=False, verbose=True
)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_no_map = inv.run(minit)
# WeightedLeastSquares Inversion
reg1 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m1, weights={"cell_weights": wr1}
)
reg2 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m2, weights={"cell_weights": wr2}
)
reg = reg1 + reg2
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
alpha0_ratio = np.r_[1, 1, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
beta_schedule = directives.BetaSchedule(coolingFactor=5.0, coolingRate=1)
targets = directives.MultiTargetMisfits(
TriggerSmall=False,
verbose=True,
)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, targets, beta_schedule, scaling_schedule],
)
mtik = inv.run(minit)
# Final Plot
fig, axes = plt.subplots(3, 4, figsize=(25, 15))
axes = axes.reshape(12)
left, width = 0.25, 0.5
bottom, height = 0.25, 0.5
right = left + width
top = bottom + height
axes[0].set_axis_off()
axes[0].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using true nonlinear\npetrophysical relationships"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[0].transAxes,
)
axes[1].plot(mesh.cell_centers_x, wires.m1 * mcluster_map, "b.-", ms=5, marker="v")
axes[1].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[1].set_title("Problem 1")
axes[1].legend(["Recovered Model", "True Model"], loc=1)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Property 1")
axes[2].plot(mesh.cell_centers_x, wires.m2 * mcluster_map, "r.-", ms=5, marker="v")
axes[2].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[2].set_title("Problem 2")
axes[2].legend(["Recovered Model", "True Model"], loc=1)
axes[2].set_xlabel("X")
axes[2].set_ylabel("Property 2")
x, y = np.mgrid[-1:1:0.01, -4:2:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
CS = axes[3].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[3].scatter(wires.m1 * mcluster_map, wires.m2 * mcluster_map, marker="v")
axes[3].set_title("Petrophysical Distribution")
CS.collections[0].set_label("")
axes[3].legend(["True Petrophysical Distribution", "Recovered model crossplot"])
axes[3].set_xlabel("Property 1")
axes[3].set_ylabel("Property 2")
axes[4].set_axis_off()
axes[4].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using a pure\nGaussian distribution"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[4].transAxes,
)
axes[5].plot(mesh.cell_centers_x, wires.m1 * mcluster_no_map, "b.-", ms=5, marker="v")
axes[5].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[5].set_title("Problem 1")
axes[5].legend(["Recovered Model", "True Model"], loc=1)
axes[5].set_xlabel("X")
axes[5].set_ylabel("Property 1")
axes[6].plot(mesh.cell_centers_x, wires.m2 * mcluster_no_map, "r.-", ms=5, marker="v")
axes[6].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[6].set_title("Problem 2")
axes[6].legend(["Recovered Model", "True Model"], loc=1)
axes[6].set_xlabel("X")
axes[6].set_ylabel("Property 2")
CSF = axes[7].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.5,
label="True Petro. Distribution",
)
CS = axes[7].contour(
x,
y,
np.exp(clfnomapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
500,
cmap="viridis",
linestyles="--",
label="Modeled Petro. Distribution",
)
axes[7].scatter(
wires.m1 * mcluster_no_map,
wires.m2 * mcluster_no_map,
marker="v",
label="Recovered model crossplot",
)
axes[7].set_title("Petrophysical Distribution")
axes[7].legend()
axes[7].set_xlabel("Property 1")
axes[7].set_ylabel("Property 2")
# Tikonov
axes[8].set_axis_off()
axes[8].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Least-Squares\n~Using a single cluster"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[8].transAxes,
)
axes[9].plot(mesh.cell_centers_x, wires.m1 * mtik, "b.-", ms=5, marker="v")
axes[9].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[9].set_title("Problem 1")
axes[9].legend(["Recovered Model", "True Model"], loc=1)
axes[9].set_xlabel("X")
axes[9].set_ylabel("Property 1")
axes[10].plot(mesh.cell_centers_x, wires.m2 * mtik, "r.-", ms=5, marker="v")
axes[10].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[10].set_title("Problem 2")
axes[10].legend(["Recovered Model", "True Model"], loc=1)
axes[10].set_xlabel("X")
axes[10].set_ylabel("Property 2")
CS = axes[11].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[11].scatter(wires.m1 * mtik, wires.m2 * mtik, marker="v")
axes[11].set_title("Petro Distribution")
CS.collections[0].set_label("")
axes[11].legend(["True Petro Distribution", "Recovered model crossplot"])
axes[11].set_xlabel("Property 1")
axes[11].set_ylabel("Property 2")
plt.subplots_adjust(wspace=0.3, hspace=0.3, top=0.85)
plt.show()
Total running time of the script: (0 minutes 41.555 seconds)
Estimated memory usage: 163 MB