Note
Go to the end to download the full example code.
Petrophysically guided inversion: Joint linear example with nonlinear relationships#
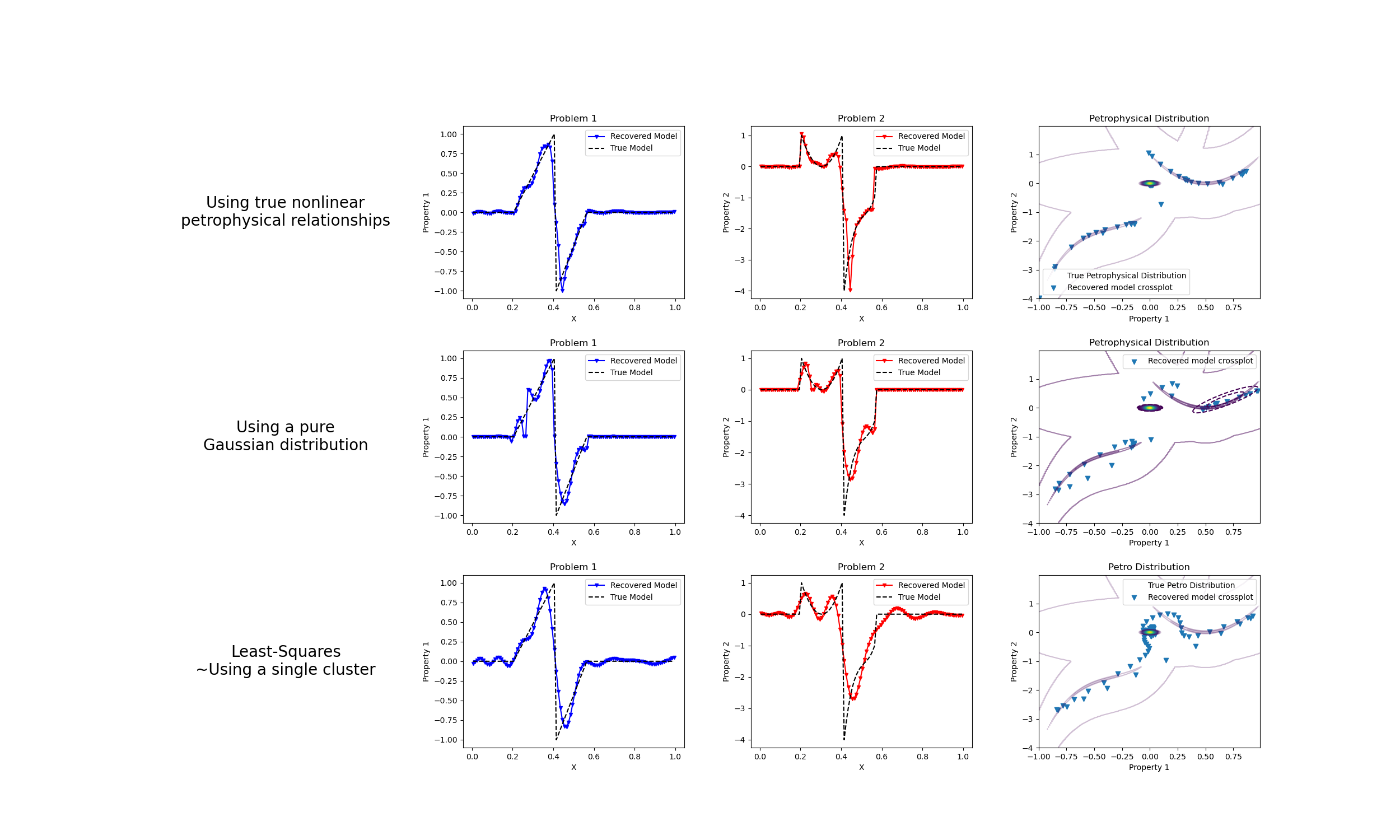
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically guided inversion. We explore it through coupling two linear problems whose respective physical properties are linked by polynomial relationships that change between rock units.
Running inversion with SimPEG v0.25.1
Alpha scales: [np.float64(3.4647755247699226), np.float64(0.0), np.float64(3.4756934888804345e-06), np.float64(0.0)]
Calculating the scaling parameter.
Scale Multipliers: [0.09295615 0.90704385]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09295615 0.90704385]
================================================= Projected GNCG =================================================
# beta phi_d phi_m f |proj(x-g)-x| LS iter_CG CG |Ax-b|/|b| CG |Ax-b| Comment
-----------------------------------------------------------------------------------------------------------------
0 1.84e+01 3.00e+05 0.00e+00 3.00e+05 0 inf inf
1 1.84e+01 1.09e+03 1.72e+02 4.26e+03 1.41e+02 0 19 6.14e-04 5.53e+03
geophys. misfits: 6711.8 (target 30.0 [False]); 510.2 (target 30.0 [False]) | smallness misfit: 3985.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [6711.8 510.2]; minimum progress targets: [240000. 240000.]
2 1.84e+01 5.92e+01 4.09e+01 8.13e+02 1.39e+02 0 100 3.12e-02 1.73e+02 Skip BFGS
geophys. misfits: 463.6 (target 30.0 [False]); 17.8 (target 30.0 [True]) | smallness misfit: 1398.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [463.6 17.8]; minimum progress targets: [5369.4 408.1]
Updating scaling for data misfits by 1.6890046719032947
New scales: [0.14755302 0.85244698]
3 1.84e+01 5.01e+01 4.14e+01 8.12e+02 9.43e+01 0 100 2.81e-02 1.12e+01 Skip BFGS
geophys. misfits: 237.1 (target 30.0 [False]); 17.8 (target 30.0 [True]) | smallness misfit: 1127.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [237.1 17.8]; minimum progress targets: [370.9 30. ]
Updating scaling for data misfits by 1.6887756659603412
New scales: [0.22619549 0.77380451]
4 1.84e+01 4.26e+01 4.24e+01 8.25e+02 7.58e+01 0 100 6.87e-03 2.25e+00
geophys. misfits: 127.4 (target 30.0 [False]); 17.8 (target 30.0 [True]) | smallness misfit: 1065.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [127.4 17.8]; minimum progress targets: [189.7 30. ]
Updating scaling for data misfits by 1.681977495668494
New scales: [0.32961 0.67039]
5 1.84e+01 3.70e+01 4.32e+01 8.33e+02 7.57e+01 0 100 8.86e-02 2.63e+01 Skip BFGS
geophys. misfits: 74.8 (target 30.0 [False]); 18.4 (target 30.0 [True]) | smallness misfit: 1016.1 (target: 200.0 [False])
Beta cooling evaluation: progress: [74.8 18.4]; minimum progress targets: [101.9 30. ]
Updating scaling for data misfits by 1.6291033890245386
New scales: [0.44474665 0.55525335]
6 1.84e+01 3.31e+01 4.37e+01 8.37e+02 7.37e+01 0 100 4.37e-02 1.06e+01 Skip BFGS
geophys. misfits: 50.1 (target 30.0 [False]); 19.5 (target 30.0 [True]) | smallness misfit: 977.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [50.1 19.5]; minimum progress targets: [59.8 30. ]
Updating scaling for data misfits by 1.5395875440868958
New scales: [0.55220773 0.44779227]
7 1.84e+01 3.06e+01 4.39e+01 8.40e+02 7.04e+01 0 100 4.06e-02 6.77e+00 Skip BFGS
geophys. misfits: 38.2 (target 30.0 [False]); 21.2 (target 30.0 [True]) | smallness misfit: 942.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [38.2 21.2]; minimum progress targets: [40. 30.]
Updating scaling for data misfits by 1.4167349732787933
New scales: [0.63597802 0.36402198]
8 1.84e+01 2.91e+01 4.41e+01 8.41e+02 6.26e+01 0 100 2.10e-01 2.29e+01 Skip BFGS
geophys. misfits: 32.5 (target 30.0 [False]); 23.4 (target 30.0 [True]) | smallness misfit: 913.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [32.5 23.4]; minimum progress targets: [30.6 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.283836193807805
New scales: [0.69164101 0.30835899]
9 9.21e+00 1.64e+01 4.50e+01 4.31e+02 8.22e+01 0 100 4.97e-02 2.09e+01
geophys. misfits: 15.7 (target 30.0 [True]); 18.0 (target 30.0 [True]) | smallness misfit: 950.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [15.7 18. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.7881818256555877
10 9.21e+00 1.76e+01 4.64e+01 4.45e+02 5.61e+01 0 100 1.50e+01 1.34e+03
geophys. misfits: 15.3 (target 30.0 [True]); 22.7 (target 30.0 [True]) | smallness misfit: 823.1 (target: 200.0 [False])
Beta cooling evaluation: progress: [15.3 22.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.9353667728917463
11 9.21e+00 1.94e+01 4.82e+01 4.63e+02 7.88e+01 0 100 6.68e-02 9.01e+01
geophys. misfits: 15.1 (target 30.0 [True]); 29.2 (target 30.0 [True]) | smallness misfit: 705.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [15.1 29.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 4.432631801816288
12 9.21e+00 2.25e+01 5.00e+01 4.84e+02 8.33e+01 0 100 7.77e-01 1.33e+02
geophys. misfits: 15.2 (target 30.0 [True]); 38.9 (target 30.0 [False]) | smallness misfit: 599.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [15.2 38.9]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.9759321577993447
New scales: [0.53164816 0.46835184]
13 4.61e+00 1.50e+01 5.14e+01 2.52e+02 9.32e+01 0 100 4.02e+00 8.65e+02
geophys. misfits: 11.7 (target 30.0 [True]); 18.7 (target 30.0 [True]) | smallness misfit: 677.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [11.7 18.7]; minimum progress targets: [30. 31.1]
Warming alpha_pgi to favor clustering: 9.208140233905636
14 4.61e+00 1.72e+01 5.74e+01 2.82e+02 9.91e+01 0 100 2.26e+00 2.00e+03
geophys. misfits: 11.9 (target 30.0 [True]); 23.2 (target 30.0 [True]) | smallness misfit: 526.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [11.9 23.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 17.526430705388897
15 4.61e+00 1.58e+01 6.63e+01 3.21e+02 1.03e+02 0 100 1.14e+00 2.31e+03
geophys. misfits: 12.2 (target 30.0 [True]); 19.9 (target 30.0 [True]) | smallness misfit: 444.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [12.2 19.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 34.81420666177654
16 4.61e+00 1.44e+01 8.26e+01 3.95e+02 1.16e+02 0 100 2.92e+00 7.14e+03
geophys. misfits: 13.4 (target 30.0 [True]); 15.6 (target 30.0 [True]) | smallness misfit: 360.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [13.4 15.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 72.47811387397323
17 4.61e+00 1.64e+01 1.06e+02 5.03e+02 1.28e+02 1 100 1.64e+00 1.20e+04
geophys. misfits: 13.1 (target 30.0 [True]); 20.1 (target 30.0 [True]) | smallness misfit: 310.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [13.1 20.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 136.8691440842503
18 4.61e+00 2.64e+01 1.45e+02 6.93e+02 1.31e+02 0 100 3.44e+00 1.62e+04
geophys. misfits: 20.0 (target 30.0 [True]); 33.6 (target 30.0 [False]) | smallness misfit: 238.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [20. 33.6]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.4976760621810685
New scales: [0.43115202 0.56884798]
19 2.30e+00 2.93e+01 1.23e+02 3.12e+02 1.23e+02 0 100 1.78e-01 2.39e+03
geophys. misfits: 20.7 (target 30.0 [True]); 35.8 (target 30.0 [False]) | smallness misfit: 163.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [20.7 35.8]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.4474180351667951
New scales: [0.34368083 0.65631917]
20 1.15e+00 2.55e+01 1.15e+02 1.57e+02 1.07e+02 1 100 1.01e+00 1.97e+03
geophys. misfits: 19.4 (target 30.0 [True]); 28.7 (target 30.0 [True]) | smallness misfit: 152.5 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [19.4 28.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 177.53922698509535
------------------------- STOP! -------------------------
1 : |fc-fOld| = 1.4825e+01 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 5.0486e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 1.0666e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.0666e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 20
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.25.1
Alpha scales: [np.float64(0.00034621110365951063), np.float64(0.0), np.float64(3.6474662483181542e-06), np.float64(0.0)]
Calculating the scaling parameter.
Scale Multipliers: [0.09295615 0.90704385]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09295615 0.90704385]
================================================= Projected GNCG =================================================
# beta phi_d phi_m f |proj(x-g)-x| LS iter_CG CG |Ax-b|/|b| CG |Ax-b| Comment
-----------------------------------------------------------------------------------------------------------------
0 1.87e+03 3.00e+05 0.00e+00 3.00e+05 0 inf inf
1 1.87e+03 6.40e+04 2.36e+01 1.08e+05 1.41e+02 0 15 3.08e-04 2.77e+03
geophys. misfits: 92126.6 (target 30.0 [False]); 61077.3 (target 30.0 [False]) | smallness misfit: 243.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [92126.6 61077.3]; minimum progress targets: [240000. 240000.]
2 1.87e+03 6.88e+01 5.52e-01 1.10e+03 1.36e+02 0 100 2.16e-01 5.77e+03 Skip BFGS
geophys. misfits: 571.5 (target 30.0 [False]); 17.2 (target 30.0 [True]) | smallness misfit: 137.6 (target: 200.0 [True])
Beta cooling evaluation: progress: [571.5 17.2]; minimum progress targets: [73701.2 48861.8]
Updating scaling for data misfits by 1.7396405660046144
New scales: [0.15130729 0.84869271]
3 1.87e+03 2.33e+01 1.34e-01 2.74e+02 1.26e+02 0 100 1.26e-02 7.88e+01 Skip BFGS
geophys. misfits: 55.0 (target 30.0 [False]); 17.6 (target 30.0 [True]) | smallness misfit: 50.4 (target: 200.0 [True])
Beta cooling evaluation: progress: [55. 17.6]; minimum progress targets: [457.2 30. ]
Updating scaling for data misfits by 1.7008706257099708
New scales: [0.23267923 0.76732077]
4 1.87e+03 1.95e+01 1.34e-01 2.69e+02 8.96e+01 0 100 5.54e-01 3.07e+02
geophys. misfits: 32.9 (target 30.0 [False]); 15.4 (target 30.0 [True]) | smallness misfit: 66.2 (target: 200.0 [True])
Beta cooling evaluation: progress: [32.9 15.4]; minimum progress targets: [44. 30.]
Updating scaling for data misfits by 1.9425302527370296
New scales: [0.37069119 0.62930881]
5 1.87e+03 1.85e+01 1.30e-01 2.62e+02 9.22e+01 0 100 1.37e-01 1.70e+02
geophys. misfits: 20.2 (target 30.0 [True]); 17.5 (target 30.0 [True]) | smallness misfit: 48.0 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [20.2 17.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.597922038981429
------------------------- STOP! -------------------------
1 : |fc-fOld| = 7.8737e+01 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 1.7740e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 9.2247e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.2247e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.25.1
Alpha scales: [np.float64(3.462979304028455e-05), np.float64(0.0), np.float64(3.4767092254362245e-05), np.float64(0.0)]
Calculating the scaling parameter.
Scale Multipliers: [0.09295615 0.90704385]
/home/vsts/work/1/s/simpeg/directives/_directives.py:334: UserWarning:
There is no PGI regularization. Smallness target is turned off (TriggerSmall flag)
Initial data misfit scales: [0.09295615 0.90704385]
================================================= Projected GNCG =================================================
# beta phi_d phi_m f |proj(x-g)-x| LS iter_CG CG |Ax-b|/|b| CG |Ax-b| Comment
-----------------------------------------------------------------------------------------------------------------
0 9.95e+05 3.00e+05 0.00e+00 3.00e+05 0 inf inf
1 9.95e+05 3.92e+04 4.42e-02 8.32e+04 1.41e+02 0 23 9.50e-04 8.55e+03
geophys. misfits: 59183.7 (target 30.0 [False]); 37179.0 (target 30.0 [False])
2 1.99e+05 4.00e+03 1.08e-01 2.55e+04 1.37e+02 0 95 9.94e-04 1.80e+01 Skip BFGS
geophys. misfits: 8196.4 (target 30.0 [False]); 3564.6 (target 30.0 [False])
3 3.98e+04 2.55e+02 1.41e-01 5.89e+03 1.30e+02 0 100 1.67e-03 8.19e+00 Skip BFGS
geophys. misfits: 537.8 (target 30.0 [False]); 226.2 (target 30.0 [False])
4 7.96e+03 2.78e+01 1.51e-01 1.23e+03 1.02e+02 0 100 1.26e-02 1.41e+01 Skip BFGS
geophys. misfits: 35.8 (target 30.0 [False]); 27.0 (target 30.0 [True])
Updating scaling for data misfits by 1.110295007313382
New scales: [0.10216133 0.89783867]
5 1.59e+03 1.30e+01 1.55e-01 2.59e+02 7.53e+01 0 100 8.16e+00 1.90e+03 Skip BFGS
geophys. misfits: 10.8 (target 30.0 [True]); 13.3 (target 30.0 [True])
All targets have been reached
------------------------- STOP! -------------------------
1 : |fc-fOld| = 9.6183e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 4.0571e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 7.5315e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 7.5315e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:302: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:309: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:353: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:360: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:367: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:412: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:419: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
import discretize as Mesh
import matplotlib.pyplot as plt
import matplotlib.lines as mlines
import numpy as np
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 30
jk = np.linspace(1.0, 59.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
m0 = np.zeros(mesh.nC)
m0[20:41] = np.linspace(0.0, 1.0, 21)
m0[41:57] = np.linspace(-1, 0.0, 16)
poly0 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[0.0, -4.0, 4.0])
poly1 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[-0.0, 3.0, 6.0, 6.0])
poly0_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, -4.0, 4.0])
poly1_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, 3.0, 6.0, 6.0])
cluster_mapping = [maps.IdentityMap(), poly0_inverse, poly1_inverse]
m1 = np.zeros(100)
m1[20:41] = 1.0 + (poly0 * np.vstack([m0[20:41], m1[20:41]]).T)[:, 1]
m1[41:57] = -1.0 + (poly1 * np.vstack([m0[41:57], m1[41:57]]).T)[:, 1]
model2d = np.vstack([m0, m1]).T
m = utils.mkvc(model2d)
clfmapping = utils.GaussianMixtureWithNonlinearRelationships(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
means_init=np.array(
[
[0, 0],
[m0[20:41].mean(), m1[20:41].mean()],
[m0[41:57].mean(), m1[41:57].mean()],
]
),
verbose=0,
verbose_interval=10,
cluster_mapping=cluster_mapping,
)
clfmapping = clfmapping.fit(model2d)
clfnomapping = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
verbose=0,
verbose_interval=10,
)
clfnomapping = clfnomapping.fit(model2d)
wires = maps.Wires(("m1", mesh.nC), ("m2", mesh.nC))
relatrive_error = 0.01
noise_floor = 0.0
prob1 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m1)
survey1 = prob1.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
prob2 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m2)
survey2 = prob2.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
dmis1 = data_misfit.L2DataMisfit(simulation=prob1, data=survey1)
dmis2 = data_misfit.L2DataMisfit(simulation=prob2, data=survey2)
dmis = dmis1 + dmis2
minit = np.zeros_like(m)
# Distance weighting
wr1 = np.sum(prob1.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr1 = wr1 / np.max(wr1)
wr2 = np.sum(prob2.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr2 = wr2 / np.max(wr2)
reg_simple = regularization.PGI(
mesh=mesh,
gmmref=clfmapping,
gmm=clfmapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=True,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
cg_maxiter=100,
cg_rtol=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[1e6, 1e4, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_map = inv.run(minit)
# Inversion with no nonlinear mapping
reg_simple_no_map = regularization.PGI(
mesh=mesh,
gmmref=clfnomapping,
gmm=clfnomapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=False,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
cg_maxiter=100,
cg_rtol=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple_no_map, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[100.0 * np.ones(2), 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(
chiSmall=1.0, TriggerSmall=True, TriggerTheta=False, verbose=True
)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_no_map = inv.run(minit)
# WeightedLeastSquares Inversion
reg1 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m1, weights={"cell_weights": wr1}
)
reg2 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m2, weights={"cell_weights": wr2}
)
reg = reg1 + reg2
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
cg_maxiter=100,
cg_rtol=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
alpha0_ratio = np.r_[1, 1, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
beta_schedule = directives.BetaSchedule(coolingFactor=5.0, coolingRate=1)
targets = directives.MultiTargetMisfits(
TriggerSmall=False,
verbose=True,
)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, targets, beta_schedule, scaling_schedule],
)
mtik = inv.run(minit)
# Final Plot
fig, axes = plt.subplots(3, 4, figsize=(25, 15))
axes = axes.reshape(12)
left, width = 0.25, 0.5
bottom, height = 0.25, 0.5
right = left + width
top = bottom + height
axes[0].set_axis_off()
axes[0].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using true nonlinear\npetrophysical relationships"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[0].transAxes,
)
axes[1].plot(mesh.cell_centers_x, wires.m1 * mcluster_map, "b.-", ms=5, marker="v")
axes[1].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[1].set_title("Problem 1")
axes[1].legend(["Recovered Model", "True Model"], loc=1)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Property 1")
axes[2].plot(mesh.cell_centers_x, wires.m2 * mcluster_map, "r.-", ms=5, marker="v")
axes[2].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[2].set_title("Problem 2")
axes[2].legend(["Recovered Model", "True Model"], loc=1)
axes[2].set_xlabel("X")
axes[2].set_ylabel("Property 2")
x, y = np.mgrid[-1:1:0.01, -4:2:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
CS = axes[3].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
cs_proxy = mlines.Line2D([], [], label="True Petrophysical Distribution")
ps = axes[3].scatter(
wires.m1 * mcluster_map,
wires.m2 * mcluster_map,
marker="v",
label="Recovered model crossplot",
)
axes[3].set_title("Petrophysical Distribution")
axes[3].legend(handles=[cs_proxy, ps])
axes[3].set_xlabel("Property 1")
axes[3].set_ylabel("Property 2")
axes[4].set_axis_off()
axes[4].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using a pure\nGaussian distribution"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[4].transAxes,
)
axes[5].plot(mesh.cell_centers_x, wires.m1 * mcluster_no_map, "b.-", ms=5, marker="v")
axes[5].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[5].set_title("Problem 1")
axes[5].legend(["Recovered Model", "True Model"], loc=1)
axes[5].set_xlabel("X")
axes[5].set_ylabel("Property 1")
axes[6].plot(mesh.cell_centers_x, wires.m2 * mcluster_no_map, "r.-", ms=5, marker="v")
axes[6].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[6].set_title("Problem 2")
axes[6].legend(["Recovered Model", "True Model"], loc=1)
axes[6].set_xlabel("X")
axes[6].set_ylabel("Property 2")
CSF = axes[7].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.5,
label="True Petro. Distribution",
)
CS = axes[7].contour(
x,
y,
np.exp(clfnomapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
500,
cmap="viridis",
linestyles="--",
)
axes[7].scatter(
wires.m1 * mcluster_no_map,
wires.m2 * mcluster_no_map,
marker="v",
label="Recovered model crossplot",
)
cs_modeled_proxy = mlines.Line2D(
[], [], linestyle="--", label="Modeled Petro. Distribution"
)
axes[7].set_title("Petrophysical Distribution")
axes[7].legend(handles=[cs_proxy, cs_modeled_proxy, ps])
axes[7].set_xlabel("Property 1")
axes[7].set_ylabel("Property 2")
# Tikonov
axes[8].set_axis_off()
axes[8].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Least-Squares\n~Using a single cluster"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[8].transAxes,
)
axes[9].plot(mesh.cell_centers_x, wires.m1 * mtik, "b.-", ms=5, marker="v")
axes[9].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[9].set_title("Problem 1")
axes[9].legend(["Recovered Model", "True Model"], loc=1)
axes[9].set_xlabel("X")
axes[9].set_ylabel("Property 1")
axes[10].plot(mesh.cell_centers_x, wires.m2 * mtik, "r.-", ms=5, marker="v")
axes[10].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[10].set_title("Problem 2")
axes[10].legend(["Recovered Model", "True Model"], loc=1)
axes[10].set_xlabel("X")
axes[10].set_ylabel("Property 2")
CS = axes[11].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[11].scatter(wires.m1 * mtik, wires.m2 * mtik, marker="v")
axes[11].set_title("Petro Distribution")
axes[11].legend(handles=[cs_proxy, ps])
axes[11].set_xlabel("Property 1")
axes[11].set_ylabel("Property 2")
plt.subplots_adjust(wspace=0.3, hspace=0.3, top=0.85)
plt.show()
Total running time of the script: (0 minutes 27.168 seconds)
Estimated memory usage: 321 MB
