Note
Go to the end to download the full example code
Magnetic inversion on a TreeMesh#
In this example, we demonstrate the use of a Magnetic Vector Inverison on 3D TreeMesh for the inversion of magnetic data.
The inverse problem uses the :class:’SimPEG.regularization.VectorAmplitude’ regularization borrowed from …
from SimPEG import (
data,
data_misfit,
directives,
maps,
inverse_problem,
optimization,
inversion,
regularization,
)
from SimPEG import utils
from SimPEG.utils import mkvc, sdiag
from discretize.utils import mesh_builder_xyz, refine_tree_xyz, active_from_xyz
from SimPEG.potential_fields import magnetics
import numpy as np
import matplotlib.pyplot as plt
# sphinx_gallery_thumbnail_number = 3
Setup#
Define the survey and model parameters
First we need to define the direction of the inducing field As a simple case, we pick a vertical inducing field of magnitude 50,000 nT.
np.random.seed(1)
# We will assume a vertical inducing field
H0 = (50000.0, 90.0, 0.0)
# Create grid of points for topography
# Lets create a simple Gaussian topo and set the active cells
[xx, yy] = np.meshgrid(np.linspace(-200, 200, 50), np.linspace(-200, 200, 50))
b = 100
A = 50
zz = A * np.exp(-0.5 * ((xx / b) ** 2.0 + (yy / b) ** 2.0))
topo = np.c_[utils.mkvc(xx), utils.mkvc(yy), utils.mkvc(zz)]
# Create an array of observation points
xr = np.linspace(-100.0, 100.0, 20)
yr = np.linspace(-100.0, 100.0, 20)
X, Y = np.meshgrid(xr, yr)
Z = A * np.exp(-0.5 * ((X / b) ** 2.0 + (Y / b) ** 2.0)) + 5
# Create a MAGsurvey
xyzLoc = np.c_[mkvc(X.T), mkvc(Y.T), mkvc(Z.T)]
rxLoc = magnetics.receivers.Point(xyzLoc)
srcField = magnetics.sources.SourceField(receiver_list=[rxLoc], parameters=H0)
survey = magnetics.survey.Survey(srcField)
Inversion Mesh#
Here, we create a TreeMesh with base cell size of 5 m.
# Create a mesh
h = [5, 5, 5]
padDist = np.ones((3, 2)) * 100
mesh = mesh_builder_xyz(
xyzLoc, h, padding_distance=padDist, depth_core=100, mesh_type="tree"
)
mesh = refine_tree_xyz(
mesh, topo, method="surface", octree_levels=[2, 6], finalize=True
)
# Define an active cells from topo
actv = active_from_xyz(mesh, topo)
nC = int(actv.sum())
/home/vsts/work/1/s/examples/03-magnetics/plot_inv_mag_MVI_VectorAmplitude.py:83: DeprecationWarning:
The surface option is deprecated as of `0.9.0` please update your code to use the `TreeMesh.refine_surface` functionality. It will be removed in a future version of discretize.
Forward modeling data#
We can now create a magnetization model and generate data.
model_azm_dip = np.zeros((mesh.nC, 2))
model_amp = np.ones(mesh.nC) * 1e-8
ind = utils.model_builder.getIndicesBlock(
np.r_[-30, -20, -10],
np.r_[30, 20, 25],
mesh.gridCC,
)[0]
model_amp[ind] = 0.05
model_azm_dip[ind, 0] = 45.0
model_azm_dip[ind, 1] = 90.0
# Remove air cells
model_azm_dip = model_azm_dip[actv, :]
model_amp = model_amp[actv]
model = sdiag(model_amp) * utils.mat_utils.dip_azimuth2cartesian(
model_azm_dip[:, 0], model_azm_dip[:, 1]
)
# Create reduced identity map
idenMap = maps.IdentityMap(nP=nC * 3)
# Create the simulation
simulation = magnetics.simulation.Simulation3DIntegral(
survey=survey, mesh=mesh, chiMap=idenMap, ind_active=actv, model_type="vector"
)
# Compute some data and add some random noise
d = simulation.dpred(mkvc(model))
std = 10 # nT
synthetic_data = d + np.random.randn(len(d)) * std
wd = np.ones(len(d)) * std
# Assign data and uncertainties to the survey
data_object = data.Data(survey, dobs=synthetic_data, standard_deviation=wd)
# Create a projection matrix for plotting later
actv_plot = maps.InjectActiveCells(mesh, actv, np.nan)
Inversion#
We can now attempt the inverse calculations.
# Create sensitivity weights from our linear forward operator
rxLoc = survey.source_field.receiver_list[0].locations
# This Mapping connects the regularizations for the three-component
# vector model
wires = maps.Wires(("p", nC), ("s", nC), ("t", nC))
m0 = np.ones(3 * nC) * 1e-4 # Starting model
# Create the regularization on the amplitude of magnetization
reg = regularization.VectorAmplitude(
mesh,
mapping=idenMap,
active_cells=actv,
reference_model_in_smooth=True,
norms=[0.0, 2.0, 2.0, 2.0],
gradient_type="total",
)
# Data misfit function
dmis = data_misfit.L2DataMisfit(simulation=simulation, data=data_object)
dmis.W = 1.0 / data_object.standard_deviation
# The optimization scheme
opt = optimization.ProjectedGNCG(
maxIter=20, lower=-10, upper=10.0, maxIterLS=20, maxIterCG=20, tolCG=1e-4
)
# The inverse problem
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# Estimate the initial beta factor
betaest = directives.BetaEstimate_ByEig(beta0_ratio=1e1)
# Add sensitivity weights
sensitivity_weights = directives.UpdateSensitivityWeights()
# Here is where the norms are applied
IRLS = directives.Update_IRLS(f_min_change=1e-3, max_irls_iterations=10, beta_tol=5e-1)
# Pre-conditioner
update_Jacobi = directives.UpdatePreconditioner()
inv = inversion.BaseInversion(
invProb, directiveList=[sensitivity_weights, IRLS, update_Jacobi, betaest]
)
# Run the inversion
mrec = inv.run(m0)
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem will set Regularization.reference_model to m0.
SimPEG.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.30e+05 5.96e+03 0.00e+00 5.96e+03 2.21e+03 0
1 6.51e+04 3.53e+03 7.39e-03 4.01e+03 1.81e+03 0
2 3.25e+04 2.34e+03 1.91e-02 2.97e+03 1.67e+03 0 Skip BFGS
3 1.63e+04 1.36e+03 3.97e-02 2.01e+03 1.56e+03 0 Skip BFGS
4 8.14e+03 6.73e+02 6.78e-02 1.22e+03 1.44e+03 0 Skip BFGS
5 4.07e+03 2.99e+02 9.72e-02 6.94e+02 1.30e+03 0 Skip BFGS
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 0.008928960443023205
6 2.03e+03 1.29e+02 2.24e-01 5.85e+02 7.99e+02 0 Skip BFGS
7 2.03e+03 1.01e+02 3.49e-01 8.10e+02 1.26e+03 0
8 2.03e+03 1.62e+02 4.57e-01 1.09e+03 1.39e+03 0
9 2.03e+03 2.48e+02 5.68e-01 1.40e+03 1.54e+03 0
10 1.34e+03 3.55e+02 5.79e-01 1.13e+03 1.23e+03 0
11 9.46e+02 3.00e+02 6.13e-01 8.80e+02 1.15e+03 0 Skip BFGS
12 9.46e+02 2.63e+02 6.02e-01 8.33e+02 1.36e+03 0 Skip BFGS
13 9.46e+02 2.76e+02 5.39e-01 7.87e+02 1.21e+03 0
14 9.46e+02 2.77e+02 4.96e-01 7.47e+02 1.14e+03 0 Skip BFGS
15 9.46e+02 2.75e+02 4.63e-01 7.14e+02 1.09e+03 0 Skip BFGS
Reach maximum number of IRLS cycles: 10
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 5.9649e+02
1 : |xc-x_last| = 2.7282e-02 <= tolX*(1+|x0|) = 1.0290e-01
0 : |proj(x-g)-x| = 1.0883e+03 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.0883e+03 <= 1e3*eps = 1.0000e-02
0 : maxIter = 20 <= iter = 16
------------------------- DONE! -------------------------
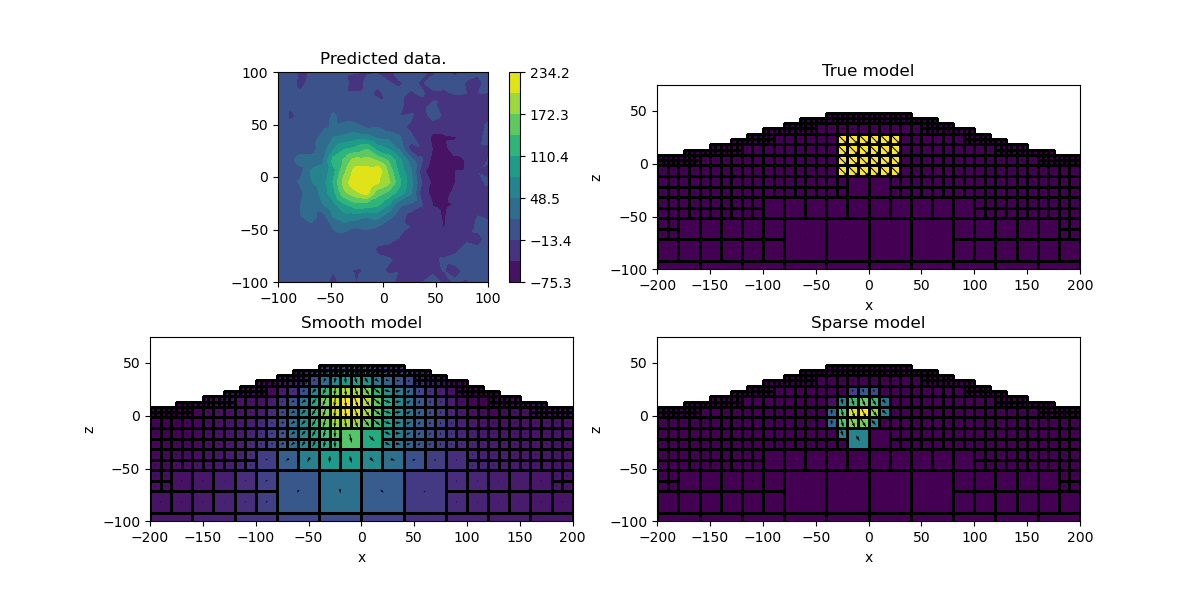
Final Plot#
Let’s compare the smooth and compact model
plt.figure(figsize=(12, 6))
ax = plt.subplot(2, 2, 1)
im = utils.plot_utils.plot2Ddata(xyzLoc, synthetic_data, ax=ax)
plt.colorbar(im[0])
ax.set_title("Predicted data.")
plt.gca().set_aspect("equal", adjustable="box")
for ii, (title, mvec) in enumerate(
[("True model", model), ("Smooth model", invProb.l2model), ("Sparse model", mrec)]
):
ax = plt.subplot(2, 2, ii + 2)
mesh.plot_slice(
actv_plot * mvec.reshape((-1, 3), order="F"),
v_type="CCv",
view="vec",
ax=ax,
normal="Y",
grid=True,
quiver_opts={
"pivot": "mid",
"scale": 8 * np.abs(mvec).max(),
"scale_units": "inches",
},
)
ax.set_xlim([-200, 200])
ax.set_ylim([-100, 75])
ax.set_title(title)
ax.set_xlabel("x")
ax.set_ylabel("z")
plt.gca().set_aspect("equal", adjustable="box")
plt.show()
print("END")
# Plot the final predicted data and the residual
# plt.figure()
# ax = plt.subplot(1, 2, 1)
# utils.plot_utils.plot2Ddata(xyzLoc, invProb.dpred, ax=ax)
# ax.set_title("Predicted data.")
# plt.gca().set_aspect("equal", adjustable="box")
#
# ax = plt.subplot(1, 2, 2)
# utils.plot_utils.plot2Ddata(xyzLoc, synthetic_data - invProb.dpred, ax=ax)
# ax.set_title("Data residual.")
# plt.gca().set_aspect("equal", adjustable="box")
END
Total running time of the script: (0 minutes 31.897 seconds)
Estimated memory usage: 137 MB