Note
Go to the end to download the full example code.
Sparse Inversion with Iteratively Re-Weighted Least-Squares#
Least-squares inversion produces smooth models which may not be an accurate representation of the true model. Here we demonstrate the basics of inverting for sparse and/or blocky models. Here, we used the iteratively reweighted least-squares approach. For this tutorial, we focus on the following:
Defining the forward problem
Defining the inverse problem (data misfit, regularization, optimization)
Defining the paramters for the IRLS algorithm
Specifying directives for the inversion
Recovering a set of model parameters which explains the observations
import numpy as np
import matplotlib.pyplot as plt
from discretize import TensorMesh
from simpeg import (
simulation,
maps,
data_misfit,
directives,
optimization,
regularization,
inverse_problem,
inversion,
)
# sphinx_gallery_thumbnail_number = 3
Defining the Model and Mapping#
Here we generate a synthetic model and a mappig which goes from the model space to the row space of our linear operator.
nParam = 100 # Number of model paramters
# A 1D mesh is used to define the row-space of the linear operator.
mesh = TensorMesh([nParam])
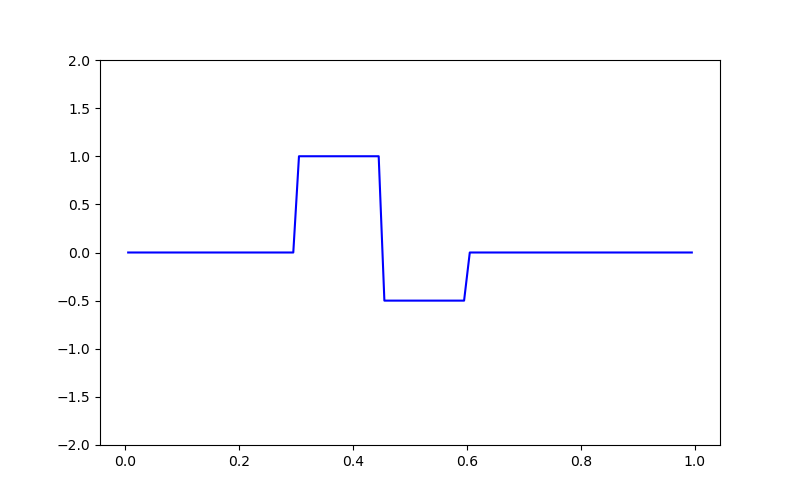
# Creating the true model
true_model = np.zeros(mesh.nC)
true_model[mesh.cell_centers_x > 0.3] = 1.0
true_model[mesh.cell_centers_x > 0.45] = -0.5
true_model[mesh.cell_centers_x > 0.6] = 0
# Mapping from the model space to the row space of the linear operator
model_map = maps.IdentityMap(mesh)
# Plotting the true model
fig = plt.figure(figsize=(8, 5))
ax = fig.add_subplot(111)
ax.plot(mesh.cell_centers_x, true_model, "b-")
ax.set_ylim([-2, 2])
(-2.0, 2.0)
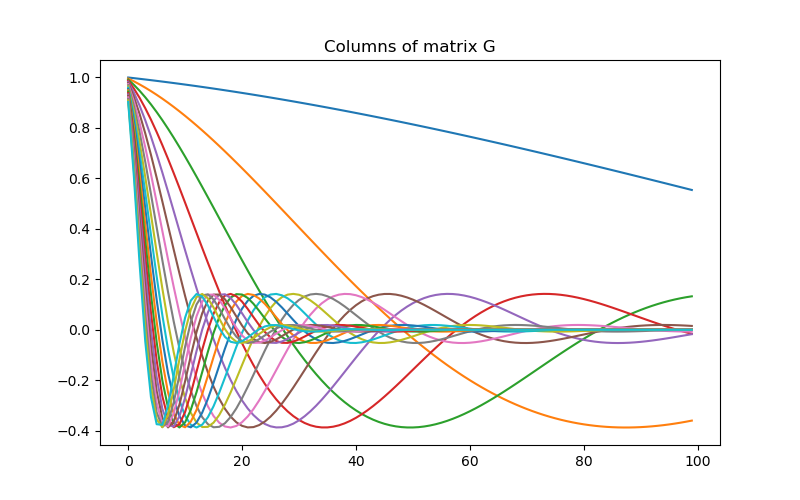
Defining the Linear Operator#
Here we define the linear operator with dimensions (nData, nParam). In practive, you may have a problem-specific linear operator which you would like to construct or load here.
# Number of data observations (rows)
nData = 20
# Create the linear operator for the tutorial. The columns of the linear operator
# represents a set of decaying and oscillating functions.
jk = np.linspace(1.0, 60.0, nData)
p = -0.25
q = 0.25
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nData, nParam))
for i in range(nData):
G[i, :] = g(i)
# Plot the columns of G
fig = plt.figure(figsize=(8, 5))
ax = fig.add_subplot(111)
for i in range(G.shape[0]):
ax.plot(G[i, :])
ax.set_title("Columns of matrix G")
Text(0.5, 1.0, 'Columns of matrix G')
Defining the Simulation#
The simulation defines the relationship between the model parameters and predicted data.
Predict Synthetic Data#
Here, we use the true model to create synthetic data which we will subsequently invert.
# Standard deviation of Gaussian noise being added
std = 0.02
np.random.seed(1)
# Create a SimPEG data object
data_obj = sim.make_synthetic_data(true_model, noise_floor=std, add_noise=True)
Define the Inverse Problem#
The inverse problem is defined by 3 things:
Data Misfit: a measure of how well our recovered model explains the field data
Regularization: constraints placed on the recovered model and a priori information
Optimization: the numerical approach used to solve the inverse problem
# Define the data misfit. Here the data misfit is the L2 norm of the weighted
# residual between the observed data and the data predicted for a given model.
# Within the data misfit, the residual between predicted and observed data are
# normalized by the data's standard deviation.
dmis = data_misfit.L2DataMisfit(simulation=sim, data=data_obj)
# Define the regularization (model objective function). Here, 'p' defines the
# the norm of the smallness term and 'q' defines the norm of the smoothness
# term.
reg = regularization.Sparse(mesh, mapping=model_map)
reg.reference_model = np.zeros(nParam)
p = 0.0
q = 0.0
reg.norms = [p, q]
# Define how the optimization problem is solved.
opt = optimization.ProjectedGNCG(
maxIter=100, lower=-2.0, upper=2.0, maxIterLS=20, maxIterCG=30, tolCG=1e-4
)
# Here we define the inverse problem that is to be solved
inv_prob = inverse_problem.BaseInvProblem(dmis, reg, opt)
Define Inversion Directives#
Here we define any directiveas that are carried out during the inversion. This includes the cooling schedule for the trade-off parameter (beta), stopping criteria for the inversion and saving inversion results at each iteration.
# Add sensitivity weights but don't update at each beta
sensitivity_weights = directives.UpdateSensitivityWeights(every_iteration=False)
# Reach target misfit for L2 solution, then use IRLS until model stops changing.
IRLS = directives.Update_IRLS(max_irls_iterations=40, minGNiter=1, f_min_change=1e-4)
# Defining a starting value for the trade-off parameter (beta) between the data
# misfit and the regularization.
starting_beta = directives.BetaEstimate_ByEig(beta0_ratio=1e0)
# Update the preconditionner
update_Jacobi = directives.UpdatePreconditioner()
# Save output at each iteration
saveDict = directives.SaveOutputEveryIteration(save_txt=False)
# Define the directives as a list
directives_list = [sensitivity_weights, IRLS, starting_beta, update_Jacobi, saveDict]
Setting a Starting Model and Running the Inversion#
To define the inversion object, we need to define the inversion problem and the set of directives. We can then run the inversion.
# Here we combine the inverse problem and the set of directives
inv = inversion.BaseInversion(inv_prob, directives_list)
# Starting model
starting_model = 1e-4 * np.ones(nParam)
# Run inversion
recovered_model = inv.run(starting_model)
Running inversion with SimPEG v0.22.1
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.70e+06 3.72e+03 1.01e-09 3.72e+03 1.99e+01 0
1 8.49e+05 1.87e+03 3.86e-04 2.20e+03 1.93e+01 0
2 4.25e+05 1.28e+03 8.90e-04 1.66e+03 1.88e+01 0 Skip BFGS
3 2.12e+05 7.48e+02 1.78e-03 1.13e+03 1.80e+01 0 Skip BFGS
4 1.06e+05 3.74e+02 3.02e-03 6.94e+02 1.65e+01 0 Skip BFGS
5 5.31e+04 1.63e+02 4.39e-03 3.96e+02 1.42e+01 0 Skip BFGS
6 2.65e+04 6.65e+01 5.64e-03 2.16e+02 1.27e+01 0 Skip BFGS
7 1.33e+04 2.90e+01 6.61e-03 1.17e+02 1.01e+01 0 Skip BFGS
Reached starting chifact with l2-norm regularization: Start IRLS steps...
irls_threshold 1.2669107150802716
8 6.63e+03 1.60e+01 1.00e-02 8.25e+01 4.05e+00 0 Skip BFGS
9 1.12e+04 1.46e+01 1.15e-02 1.43e+02 1.69e+01 0
10 8.79e+03 2.43e+01 1.15e-02 1.25e+02 2.91e+00 0
11 6.86e+03 2.46e+01 1.23e-02 1.09e+02 3.25e+00 0 Skip BFGS
12 5.45e+03 2.38e+01 1.28e-02 9.35e+01 4.40e+00 0 Skip BFGS
13 4.47e+03 2.24e+01 1.29e-02 8.01e+01 4.58e+00 0 Skip BFGS
14 4.47e+03 2.07e+01 1.26e-02 7.69e+01 6.10e+00 0
15 4.47e+03 2.07e+01 1.17e-02 7.30e+01 6.39e+00 0
16 4.47e+03 2.06e+01 1.07e-02 6.84e+01 6.50e+00 0
17 4.47e+03 2.02e+01 9.64e-03 6.33e+01 6.84e+00 0
18 4.47e+03 1.97e+01 8.56e-03 5.79e+01 7.31e+00 0
19 4.47e+03 1.89e+01 7.41e-03 5.21e+01 7.79e+00 0
20 6.97e+03 1.79e+01 6.47e-03 6.30e+01 1.50e+01 0
21 6.97e+03 2.03e+01 5.31e-03 5.73e+01 1.04e+01 0
22 6.97e+03 1.98e+01 4.57e-03 5.17e+01 1.03e+01 0 Skip BFGS
23 6.97e+03 1.90e+01 4.11e-03 4.76e+01 1.13e+01 0
24 6.97e+03 1.84e+01 3.75e-03 4.45e+01 1.19e+01 0
25 6.97e+03 1.87e+01 3.20e-03 4.10e+01 1.77e+01 0
26 1.09e+04 1.77e+01 2.71e-03 4.72e+01 1.70e+01 0
27 1.09e+04 1.87e+01 2.17e-03 4.24e+01 1.35e+01 0
28 1.09e+04 1.82e+01 1.80e-03 3.79e+01 1.26e+01 0
29 1.71e+04 1.77e+01 1.50e-03 4.34e+01 1.87e+01 0
30 1.71e+04 1.85e+01 1.19e-03 3.88e+01 1.41e+01 0
31 1.71e+04 1.85e+01 1.02e-03 3.59e+01 1.64e+01 1 Skip BFGS
32 1.71e+04 1.89e+01 8.52e-04 3.34e+01 1.35e+01 0
33 1.71e+04 1.92e+01 7.23e-04 3.15e+01 1.32e+01 0 Skip BFGS
34 1.71e+04 1.95e+01 6.14e-04 3.00e+01 1.37e+01 0
35 1.71e+04 1.98e+01 5.23e-04 2.87e+01 1.39e+01 0
36 1.71e+04 2.01e+01 4.46e-04 2.77e+01 1.41e+01 0
37 1.71e+04 2.03e+01 3.80e-04 2.68e+01 1.42e+01 0
38 1.71e+04 2.05e+01 3.17e-04 2.59e+01 1.40e+01 0
39 1.71e+04 2.05e+01 2.66e-04 2.51e+01 1.40e+01 0
40 1.71e+04 2.06e+01 2.26e-04 2.44e+01 1.38e+01 0
41 1.71e+04 2.06e+01 1.89e-04 2.38e+01 1.37e+01 0
42 1.71e+04 2.06e+01 1.70e-04 2.35e+01 1.68e+01 2
43 1.71e+04 2.07e+01 1.53e-04 2.33e+01 1.76e+01 2 Skip BFGS
44 1.71e+04 2.11e+01 1.17e-04 2.31e+01 1.72e+01 0
45 1.71e+04 2.11e+01 9.65e-05 2.27e+01 1.36e+01 0
46 1.71e+04 2.11e+01 8.97e-05 2.26e+01 1.64e+01 8 Skip BFGS
47 1.71e+04 2.11e+01 8.29e-05 2.25e+01 1.78e+01 3
Reach maximum number of IRLS cycles: 40
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.7168e+02
1 : |xc-x_last| = 4.0540e-02 <= tolX*(1+|x0|) = 1.0010e-01
0 : |proj(x-g)-x| = 1.7789e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.7789e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 100 <= iter = 48
------------------------- DONE! -------------------------
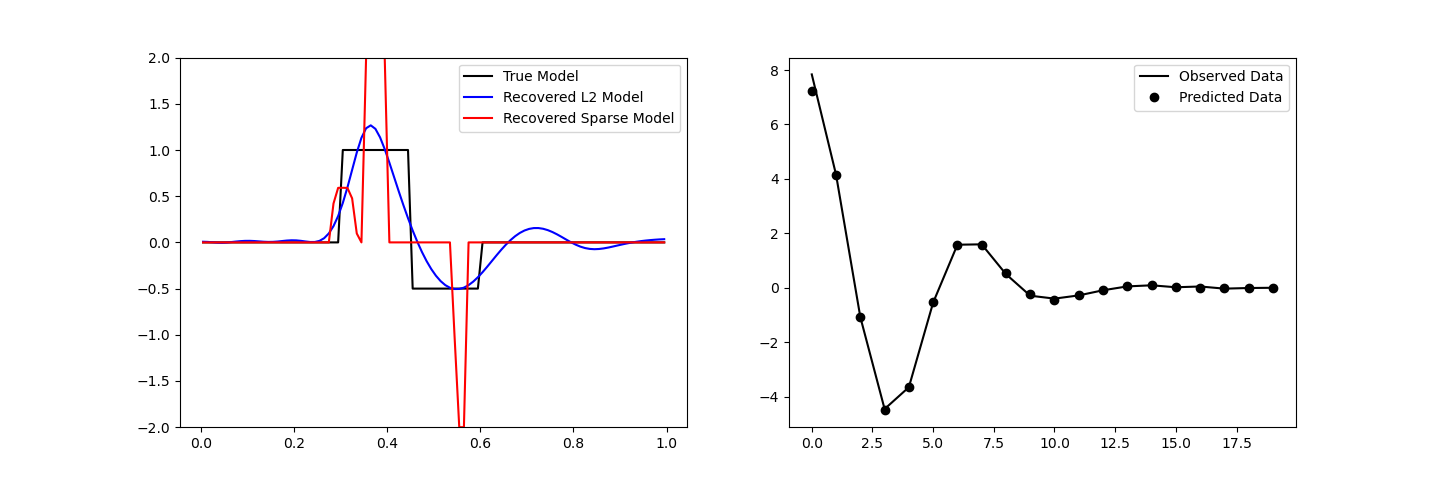
Plotting Results#
fig, ax = plt.subplots(1, 2, figsize=(12 * 1.2, 4 * 1.2))
# True versus recovered model
ax[0].plot(mesh.cell_centers_x, true_model, "k-")
ax[0].plot(mesh.cell_centers_x, inv_prob.l2model, "b-")
ax[0].plot(mesh.cell_centers_x, recovered_model, "r-")
ax[0].legend(("True Model", "Recovered L2 Model", "Recovered Sparse Model"))
ax[0].set_ylim([-2, 2])
# Observed versus predicted data
ax[1].plot(data_obj.dobs, "k-")
ax[1].plot(inv_prob.dpred, "ko")
ax[1].legend(("Observed Data", "Predicted Data"))
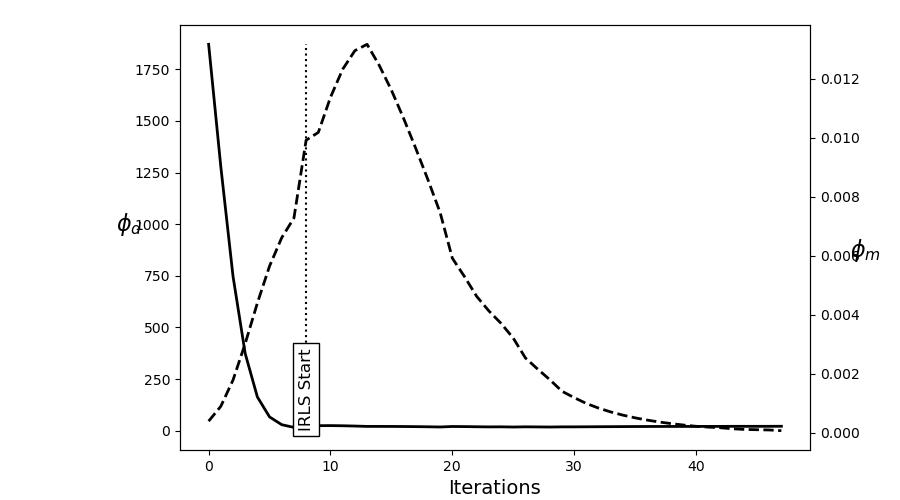
# Plot convergence
fig = plt.figure(figsize=(9, 5))
ax = fig.add_axes([0.2, 0.1, 0.7, 0.85])
ax.plot(saveDict.phi_d, "k", lw=2)
twin = ax.twinx()
twin.plot(saveDict.phi_m, "k--", lw=2)
ax.plot(np.r_[IRLS.iterStart, IRLS.iterStart], np.r_[0, np.max(saveDict.phi_d)], "k:")
ax.text(
IRLS.iterStart,
0.0,
"IRLS Start",
va="bottom",
ha="center",
rotation="vertical",
size=12,
bbox={"facecolor": "white"},
)
ax.set_ylabel(r"$\phi_d$", size=16, rotation=0)
ax.set_xlabel("Iterations", size=14)
twin.set_ylabel(r"$\phi_m$", size=16, rotation=0)
Text(865.1527777777777, 0.5, '$\\phi_m$')
Total running time of the script: (0 minutes 27.241 seconds)
Estimated memory usage: 9 MB