Note
Go to the end to download the full example code.
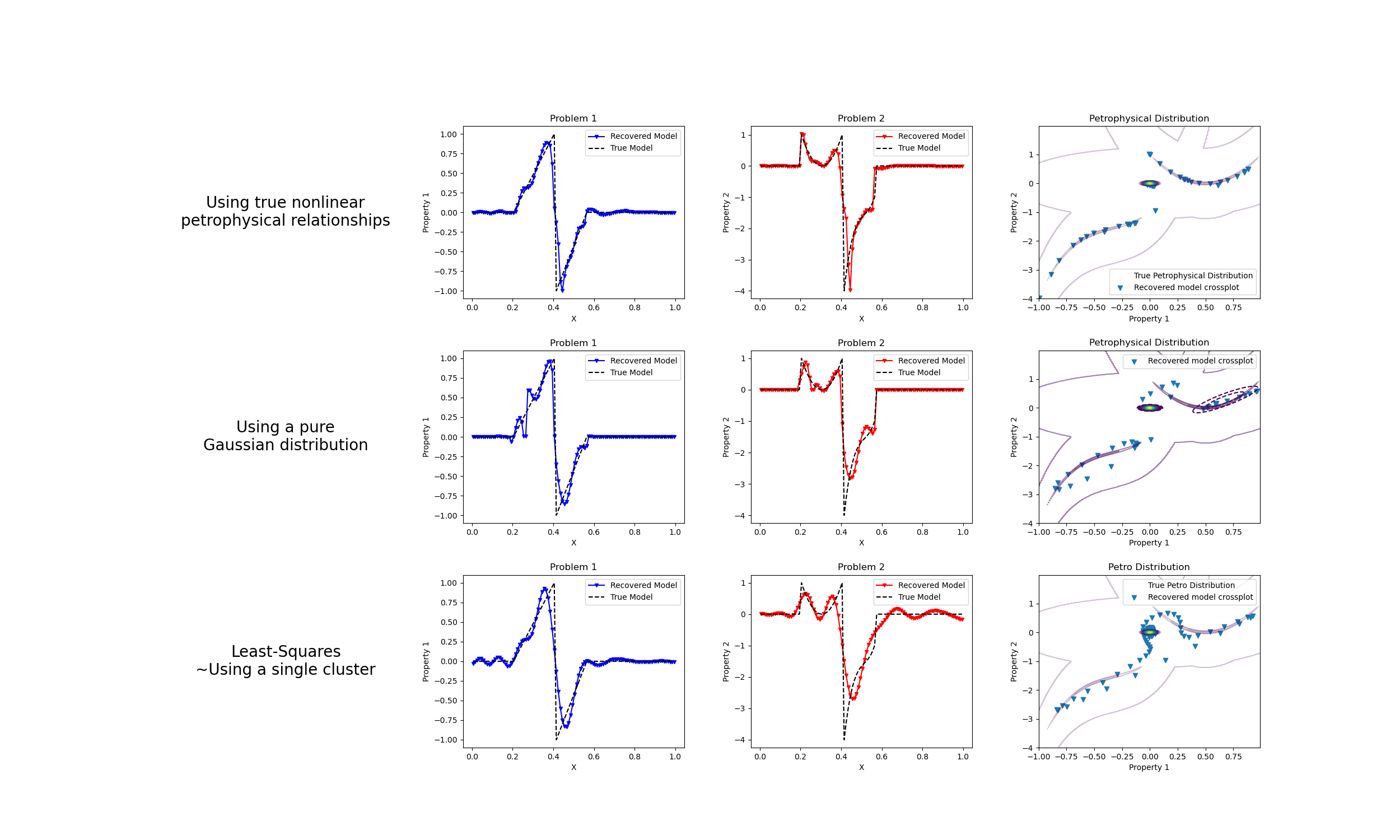
Petrophysically guided inversion: Joint linear example with nonlinear relationships#
We do a comparison between the classic least-squares inversion and our formulation of a petrophysically guided inversion. We explore it through coupling two linear problems whose respective physical properties are linked by polynomial relationships that change between rock units.
Running inversion with SimPEG v0.22.0
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.5111974374988844, 0.0, 3.4802923651593253e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09429751 0.90570249]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09429751 0.90570249]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.91e+01 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 1079.1 (target 30.0 [False]); 83.7 (target 30.0 [False]) | smallness misfit: 2929.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [1079.1 83.7]; minimum progress targets: [240000. 240000.]
1 1.91e+01 1.78e+02 4.14e+01 9.69e+02 8.21e+01 0
geophys. misfits: 482.7 (target 30.0 [False]); 28.2 (target 30.0 [True]) | smallness misfit: 1350.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [482.7 28.2]; minimum progress targets: [863.3 67. ]
Updating scaling for data misfits by 1.0620202907137457
New scales: [0.09956358 0.90043642]
2 1.91e+01 7.35e+01 4.04e+01 8.45e+02 3.73e+01 0 Skip BFGS
geophys. misfits: 439.6 (target 30.0 [False]); 28.1 (target 30.0 [True]) | smallness misfit: 1319.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [439.6 28.1]; minimum progress targets: [386.2 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.0663187291080125
New scales: [0.1054701 0.8945299]
3 9.56e+00 7.15e+01 4.06e+01 4.59e+02 8.41e+01 0 Skip BFGS
geophys. misfits: 148.9 (target 30.0 [False]); 22.7 (target 30.0 [True]) | smallness misfit: 1358.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [148.9 22.7]; minimum progress targets: [351.7 30. ]
Updating scaling for data misfits by 1.3196861608421822
New scales: [0.13464748 0.86535252]
4 9.56e+00 3.97e+01 4.30e+01 4.51e+02 5.69e+01 0
geophys. misfits: 118.6 (target 30.0 [False]); 23.5 (target 30.0 [True]) | smallness misfit: 1290.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [118.6 23.5]; minimum progress targets: [119.1 30. ]
Updating scaling for data misfits by 1.2791257490712276
New scales: [0.16599247 0.83400753]
5 9.56e+00 3.92e+01 4.33e+01 4.53e+02 6.36e+01 0 Skip BFGS
geophys. misfits: 81.1 (target 30.0 [False]); 23.1 (target 30.0 [True]) | smallness misfit: 1275.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [81.1 23.1]; minimum progress targets: [94.9 30. ]
Updating scaling for data misfits by 1.298685061013985
New scales: [0.20538886 0.79461114]
6 9.56e+00 3.50e+01 4.39e+01 4.55e+02 5.61e+01 0
geophys. misfits: 63.3 (target 30.0 [False]); 23.2 (target 30.0 [True]) | smallness misfit: 1244.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [63.3 23.2]; minimum progress targets: [64.9 30. ]
Updating scaling for data misfits by 1.2926615014373246
New scales: [0.25044422 0.74955578]
7 9.56e+00 3.33e+01 4.42e+01 4.56e+02 5.75e+01 0 Skip BFGS
geophys. misfits: 53.0 (target 30.0 [False]); 23.7 (target 30.0 [True]) | smallness misfit: 1219.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [53. 23.7]; minimum progress targets: [50.6 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.263540875307549
New scales: [0.2968535 0.7031465]
8 4.78e+00 3.24e+01 4.44e+01 2.45e+02 7.61e+01 0
geophys. misfits: 28.9 (target 30.0 [True]); 20.9 (target 30.0 [True]) | smallness misfit: 1315.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [28.9 20.9]; minimum progress targets: [42.4 30. ]
Warming alpha_pgi to favor clustering: 1.2344826360406898
9 4.78e+00 2.33e+01 4.63e+01 2.45e+02 7.09e+01 0
geophys. misfits: 28.2 (target 30.0 [True]); 21.4 (target 30.0 [True]) | smallness misfit: 1242.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [28.2 21.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.5218665825639803
10 4.78e+00 2.34e+01 4.69e+01 2.48e+02 6.06e+01 0
geophys. misfits: 26.3 (target 30.0 [True]); 21.9 (target 30.0 [True]) | smallness misfit: 1152.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.3 21.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 1.908281988237571
11 4.78e+00 2.32e+01 4.78e+01 2.52e+02 4.72e+01 0
geophys. misfits: 25.4 (target 30.0 [True]); 22.5 (target 30.0 [True]) | smallness misfit: 1073.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.4 22.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.400424867767163
12 4.78e+00 2.33e+01 4.89e+01 2.57e+02 3.17e+01 0
geophys. misfits: 24.9 (target 30.0 [True]); 23.2 (target 30.0 [True]) | smallness misfit: 987.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.9 23.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 2.9976387400152387
13 4.78e+00 2.37e+01 5.00e+01 2.63e+02 5.81e+01 0
geophys. misfits: 24.5 (target 30.0 [True]); 24.1 (target 30.0 [True]) | smallness misfit: 912.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.5 24.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 3.6965455513930157
14 4.78e+00 2.42e+01 5.12e+01 2.69e+02 6.56e+01 0
geophys. misfits: 24.9 (target 30.0 [True]); 25.1 (target 30.0 [True]) | smallness misfit: 838.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.9 25.1]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 4.4339769943829594
15 4.78e+00 2.50e+01 5.23e+01 2.75e+02 7.87e+01 0
geophys. misfits: 24.5 (target 30.0 [True]); 25.7 (target 30.0 [True]) | smallness misfit: 782.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.5 25.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 5.305715646440647
16 4.78e+00 2.53e+01 5.37e+01 2.82e+02 7.23e+01 0
geophys. misfits: 24.8 (target 30.0 [True]); 26.8 (target 30.0 [True]) | smallness misfit: 702.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.8 26.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 6.174049240549029
17 4.78e+00 2.62e+01 5.47e+01 2.87e+02 7.78e+01 0
geophys. misfits: 25.0 (target 30.0 [True]); 27.5 (target 30.0 [True]) | smallness misfit: 651.7 (target: 200.0 [False])
Beta cooling evaluation: progress: [25. 27.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 7.077074939554095
18 4.78e+00 2.67e+01 5.58e+01 2.93e+02 5.73e+01 0 Skip BFGS
geophys. misfits: 25.4 (target 30.0 [True]); 28.2 (target 30.0 [True]) | smallness misfit: 607.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.4 28.2]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 7.954968826192312
19 4.78e+00 2.73e+01 5.68e+01 2.99e+02 6.48e+01 0 Skip BFGS
geophys. misfits: 25.6 (target 30.0 [True]); 28.8 (target 30.0 [True]) | smallness misfit: 569.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [25.6 28.8]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 8.798348914444245
20 4.78e+00 2.79e+01 5.77e+01 3.03e+02 6.53e+01 0 Skip BFGS
geophys. misfits: 26.2 (target 30.0 [True]); 29.4 (target 30.0 [True]) | smallness misfit: 538.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.2 29.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 9.533408118000374
21 4.78e+00 2.84e+01 5.84e+01 3.07e+02 7.85e+01 0 Skip BFGS
geophys. misfits: 26.4 (target 30.0 [True]); 30.1 (target 30.0 [False]) | smallness misfit: 515.1 (target: 200.0 [False])
Beta cooling evaluation: progress: [26.4 30.1]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.1375763963523058
New scales: [0.27066988 0.72933012]
22 2.39e+00 2.91e+01 5.82e+01 1.68e+02 8.85e+01 0
geophys. misfits: 23.7 (target 30.0 [True]); 24.9 (target 30.0 [True]) | smallness misfit: 564.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.7 24.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 11.784831908155274
23 2.39e+00 2.46e+01 6.23e+01 1.73e+02 8.44e+01 0
geophys. misfits: 23.1 (target 30.0 [True]); 24.3 (target 30.0 [True]) | smallness misfit: 518.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.1 24.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 14.931368471700454
24 2.39e+00 2.40e+01 6.58e+01 1.81e+02 9.30e+01 0
geophys. misfits: 21.8 (target 30.0 [True]); 23.9 (target 30.0 [True]) | smallness misfit: 491.9 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.8 23.9]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 19.650374200956737
25 2.39e+00 2.33e+01 7.09e+01 1.93e+02 1.02e+02 0
geophys. misfits: 21.8 (target 30.0 [True]); 22.7 (target 30.0 [True]) | smallness misfit: 443.6 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.8 22.7]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 26.510066720861296
26 2.39e+00 2.25e+01 7.74e+01 2.08e+02 1.03e+02 0
geophys. misfits: 21.3 (target 30.0 [True]); 22.4 (target 30.0 [True]) | smallness misfit: 415.8 (target: 200.0 [False])
Beta cooling evaluation: progress: [21.3 22.4]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 36.37715172737612
27 2.39e+00 2.21e+01 8.60e+01 2.28e+02 1.13e+02 0
geophys. misfits: 23.2 (target 30.0 [True]); 22.3 (target 30.0 [True]) | smallness misfit: 360.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [23.2 22.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 48.00309361234333
28 2.39e+00 2.25e+01 9.42e+01 2.47e+02 1.14e+02 0
geophys. misfits: 22.5 (target 30.0 [True]); 22.6 (target 30.0 [True]) | smallness misfit: 351.5 (target: 200.0 [False])
Beta cooling evaluation: progress: [22.5 22.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 63.879518397111084
29 2.39e+00 2.26e+01 1.05e+02 2.74e+02 1.19e+02 0
geophys. misfits: 27.4 (target 30.0 [True]); 22.3 (target 30.0 [True]) | smallness misfit: 306.3 (target: 200.0 [False])
Beta cooling evaluation: progress: [27.4 22.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 78.02898057143261
30 2.39e+00 2.36e+01 1.12e+02 2.92e+02 1.27e+02 0
geophys. misfits: 24.6 (target 30.0 [True]); 23.0 (target 30.0 [True]) | smallness misfit: 278.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [24.6 23. ]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 98.43114270782824
31 2.39e+00 2.34e+01 1.21e+02 3.13e+02 1.19e+02 0
geophys. misfits: 36.6 (target 30.0 [False]); 25.6 (target 30.0 [True]) | smallness misfit: 226.2 (target: 200.0 [False])
Beta cooling evaluation: progress: [36.6 25.6]; minimum progress targets: [30. 30.]
Decreasing beta to counter data misfit increase.
Updating scaling for data misfits by 1.1732718933185409
New scales: [0.30334275 0.69665725]
32 1.19e+00 2.89e+01 1.16e+02 1.67e+02 1.18e+02 0
geophys. misfits: 27.1 (target 30.0 [True]); 25.5 (target 30.0 [True]) | smallness misfit: 213.4 (target: 200.0 [False])
Beta cooling evaluation: progress: [27.1 25.5]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 112.27045537462944
33 1.19e+00 2.60e+01 1.21e+02 1.71e+02 1.14e+02 0
geophys. misfits: 28.2 (target 30.0 [True]); 24.3 (target 30.0 [True]) | smallness misfit: 211.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [28.2 24.3]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 129.08852518299622
34 1.19e+00 2.55e+01 1.28e+02 1.78e+02 1.10e+02 0
geophys. misfits: 29.2 (target 30.0 [True]); 25.6 (target 30.0 [True]) | smallness misfit: 194.5 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [29.2 25.6]; minimum progress targets: [30. 30.]
Warming alpha_pgi to favor clustering: 142.12597606212722
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 5.0874e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 1.1035e+02 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 1.1035e+02 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 35
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.0
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [0.00035290147269136, 0.0, 3.5122265027259625e-06, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09429751 0.90570249]
<class 'simpeg.regularization.pgi.PGIsmallness'>
Initial data misfit scales: [0.09429751 0.90570249]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.91e+03 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 88746.2 (target 30.0 [False]); 62261.7 (target 30.0 [False]) | smallness misfit: 272.0 (target: 200.0 [False])
Beta cooling evaluation: progress: [88746.2 62261.7]; minimum progress targets: [240000. 240000.]
1 1.91e+03 6.48e+04 6.57e-01 6.60e+04 9.27e+01 0
geophys. misfits: 642.5 (target 30.0 [False]); 24.1 (target 30.0 [True]) | smallness misfit: 112.7 (target: 200.0 [True])
Beta cooling evaluation: progress: [642.5 24.1]; minimum progress targets: [70996.9 49809.3]
Updating scaling for data misfits by 1.2471548335361147
New scales: [0.11492514 0.88507486]
2 1.91e+03 9.51e+01 3.23e-01 7.14e+02 8.44e+01 0 Skip BFGS
geophys. misfits: 107.4 (target 30.0 [False]); 22.3 (target 30.0 [True]) | smallness misfit: 61.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [107.4 22.3]; minimum progress targets: [514. 30.]
Updating scaling for data misfits by 1.3427652544537596
New scales: [0.14846894 0.85153106]
3 1.91e+03 3.50e+01 1.53e-01 3.28e+02 1.05e+02 0 Skip BFGS
geophys. misfits: 76.3 (target 30.0 [False]); 21.4 (target 30.0 [True]) | smallness misfit: 46.6 (target: 200.0 [True])
Beta cooling evaluation: progress: [76.3 21.4]; minimum progress targets: [85.9 30. ]
Updating scaling for data misfits by 1.3995646935052177
New scales: [0.19615537 0.80384463]
4 1.91e+03 3.22e+01 1.27e-01 2.75e+02 7.57e+01 0
geophys. misfits: 56.0 (target 30.0 [False]); 22.0 (target 30.0 [True]) | smallness misfit: 47.4 (target: 200.0 [True])
Beta cooling evaluation: progress: [56. 22.]; minimum progress targets: [61. 30.]
Updating scaling for data misfits by 1.3642761578592137
New scales: [0.24976333 0.75023667]
5 1.91e+03 3.05e+01 1.29e-01 2.76e+02 7.88e+01 0 Skip BFGS
geophys. misfits: 44.7 (target 30.0 [False]); 22.5 (target 30.0 [True]) | smallness misfit: 47.9 (target: 200.0 [True])
Beta cooling evaluation: progress: [44.7 22.5]; minimum progress targets: [44.8 30. ]
Updating scaling for data misfits by 1.3358205199459254
New scales: [0.30782035 0.69217965]
6 1.91e+03 2.93e+01 1.30e-01 2.77e+02 7.89e+01 0 Skip BFGS
geophys. misfits: 38.0 (target 30.0 [False]); 23.0 (target 30.0 [True]) | smallness misfit: 48.3 (target: 200.0 [True])
Beta cooling evaluation: progress: [38. 23.]; minimum progress targets: [35.7 30. ]
Decreasing beta to counter data misfit decrase plateau.
Updating scaling for data misfits by 1.3017109355198204
New scales: [0.36664205 0.63335795]
7 9.57e+02 2.85e+01 1.30e-01 1.53e+02 9.91e+01 0 Skip BFGS
geophys. misfits: 26.5 (target 30.0 [True]); 20.3 (target 30.0 [True]) | smallness misfit: 50.2 (target: 200.0 [True])
All targets have been reached
Beta cooling evaluation: progress: [26.5 20.3]; minimum progress targets: [30.4 30. ]
Warming alpha_pgi to favor clustering: 1.3051451777497802
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 1.5127e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 9.9134e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 9.9134e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 8
------------------------- DONE! -------------------------
Running inversion with SimPEG v0.22.0
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem will set Regularization.reference_model to m0.
simpeg.InvProblem is setting bfgsH0 to the inverse of the eval2Deriv.
***Done using the default solver Pardiso and no solver_opts.***
Alpha scales: [3.494765446396261e-05, 0.0, 3.50340917582425e-05, 0.0]
Calculating the scaling parameter.
Scale Multipliers: [0.09429751 0.90570249]
/home/vsts/work/1/s/simpeg/directives/directives.py:334: UserWarning:
There is no PGI regularization. Smallness target is turned off (TriggerSmall flag)
Initial data misfit scales: [0.09429751 0.90570249]
model has any nan: 0
=============================== Projected GNCG ===============================
# beta phi_d phi_m f |proj(x-g)-x| LS Comment
-----------------------------------------------------------------------------
x0 has any nan: 0
0 1.03e+06 3.00e+05 0.00e+00 3.00e+05 1.41e+02 0
geophys. misfits: 57071.4 (target 30.0 [False]); 35672.8 (target 30.0 [False])
1 2.07e+05 3.77e+04 4.30e-02 4.66e+04 1.38e+02 0
geophys. misfits: 8548.9 (target 30.0 [False]); 3856.8 (target 30.0 [False])
2 4.14e+04 4.30e+03 1.07e-01 8.73e+03 1.30e+02 0 Skip BFGS
geophys. misfits: 574.4 (target 30.0 [False]); 258.8 (target 30.0 [False])
3 8.27e+03 2.89e+02 1.41e-01 1.46e+03 1.02e+02 0 Skip BFGS
geophys. misfits: 45.6 (target 30.0 [False]); 37.2 (target 30.0 [False])
4 1.65e+03 3.80e+01 1.52e-01 2.90e+02 8.81e+01 0 Skip BFGS
geophys. misfits: 21.2 (target 30.0 [True]); 19.8 (target 30.0 [True])
All targets have been reached
------------------------- STOP! -------------------------
1 : |fc-fOld| = 0.0000e+00 <= tolF*(1+|f0|) = 3.0000e+04
0 : |xc-x_last| = 4.9534e-01 <= tolX*(1+|x0|) = 1.0000e-06
0 : |proj(x-g)-x| = 8.8080e+01 <= tolG = 1.0000e-01
0 : |proj(x-g)-x| = 8.8080e+01 <= 1e3*eps = 1.0000e-02
0 : maxIter = 50 <= iter = 5
------------------------- DONE! -------------------------
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:301: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:308: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:346: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:353: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:360: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:368: UserWarning:
The following kwargs were not used by contour: 'label'
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:402: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "b.-" (-> marker='.'). The keyword argument will take precedence.
/home/vsts/work/1/s/examples/10-pgi/plot_inv_1_PGI_Linear_1D_joint_WithRelationships.py:409: UserWarning:
marker is redundantly defined by the 'marker' keyword argument and the fmt string "r.-" (-> marker='.'). The keyword argument will take precedence.
import discretize as Mesh
import matplotlib.pyplot as plt
import numpy as np
from simpeg import (
data_misfit,
directives,
inverse_problem,
inversion,
maps,
optimization,
regularization,
simulation,
utils,
)
# Random seed for reproductibility
np.random.seed(1)
# Mesh
N = 100
mesh = Mesh.TensorMesh([N])
# Survey design parameters
nk = 30
jk = np.linspace(1.0, 59.0, nk)
p = -0.25
q = 0.25
# Physics
def g(k):
return np.exp(p * jk[k] * mesh.cell_centers_x) * np.cos(
np.pi * q * jk[k] * mesh.cell_centers_x
)
G = np.empty((nk, mesh.nC))
for i in range(nk):
G[i, :] = g(i)
m0 = np.zeros(mesh.nC)
m0[20:41] = np.linspace(0.0, 1.0, 21)
m0[41:57] = np.linspace(-1, 0.0, 16)
poly0 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[0.0, -4.0, 4.0])
poly1 = maps.PolynomialPetroClusterMap(coeffyx=np.r_[-0.0, 3.0, 6.0, 6.0])
poly0_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, -4.0, 4.0])
poly1_inverse = maps.PolynomialPetroClusterMap(coeffyx=-np.r_[0.0, 3.0, 6.0, 6.0])
cluster_mapping = [maps.IdentityMap(), poly0_inverse, poly1_inverse]
m1 = np.zeros(100)
m1[20:41] = 1.0 + (poly0 * np.vstack([m0[20:41], m1[20:41]]).T)[:, 1]
m1[41:57] = -1.0 + (poly1 * np.vstack([m0[41:57], m1[41:57]]).T)[:, 1]
model2d = np.vstack([m0, m1]).T
m = utils.mkvc(model2d)
clfmapping = utils.GaussianMixtureWithNonlinearRelationships(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
means_init=np.array(
[
[0, 0],
[m0[20:41].mean(), m1[20:41].mean()],
[m0[41:57].mean(), m1[41:57].mean()],
]
),
verbose=0,
verbose_interval=10,
cluster_mapping=cluster_mapping,
)
clfmapping = clfmapping.fit(model2d)
clfnomapping = utils.WeightedGaussianMixture(
mesh=mesh,
n_components=3,
covariance_type="full",
tol=1e-8,
reg_covar=1e-3,
max_iter=1000,
n_init=100,
init_params="kmeans",
random_state=None,
warm_start=False,
verbose=0,
verbose_interval=10,
)
clfnomapping = clfnomapping.fit(model2d)
wires = maps.Wires(("m1", mesh.nC), ("m2", mesh.nC))
relatrive_error = 0.01
noise_floor = 0.0
prob1 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m1)
survey1 = prob1.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
prob2 = simulation.LinearSimulation(mesh, G=G, model_map=wires.m2)
survey2 = prob2.make_synthetic_data(
m, relative_error=relatrive_error, noise_floor=noise_floor, add_noise=True
)
dmis1 = data_misfit.L2DataMisfit(simulation=prob1, data=survey1)
dmis2 = data_misfit.L2DataMisfit(simulation=prob2, data=survey2)
dmis = dmis1 + dmis2
minit = np.zeros_like(m)
# Distance weighting
wr1 = np.sum(prob1.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr1 = wr1 / np.max(wr1)
wr2 = np.sum(prob2.G**2.0, axis=0) ** 0.5 / mesh.cell_volumes
wr2 = wr2 / np.max(wr2)
reg_simple = regularization.PGI(
mesh=mesh,
gmmref=clfmapping,
gmm=clfmapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=True,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[1e6, 1e4, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(verbose=True)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_map = inv.run(minit)
# Inversion with no nonlinear mapping
reg_simple_no_map = regularization.PGI(
mesh=mesh,
gmmref=clfnomapping,
gmm=clfnomapping,
approx_gradient=True,
wiresmap=wires,
non_linear_relationships=False,
weights_list=[wr1, wr2],
)
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg_simple_no_map, opt)
# directives
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
alpha0_ratio = np.r_[100.0 * np.ones(2), 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
betaIt = directives.PGI_BetaAlphaSchedule(
verbose=True,
coolingFactor=2.0,
progress=0.2,
)
targets = directives.MultiTargetMisfits(
chiSmall=1.0, TriggerSmall=True, TriggerTheta=False, verbose=True
)
petrodir = directives.PGI_UpdateParameters(update_gmm=False)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, petrodir, targets, betaIt, scaling_schedule],
)
mcluster_no_map = inv.run(minit)
# WeightedLeastSquares Inversion
reg1 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m1, weights={"cell_weights": wr1}
)
reg2 = regularization.WeightedLeastSquares(
mesh, alpha_s=1.0, alpha_x=1.0, mapping=wires.m2, weights={"cell_weights": wr2}
)
reg = reg1 + reg2
opt = optimization.ProjectedGNCG(
maxIter=50,
tolX=1e-6,
maxIterCG=100,
tolCG=1e-3,
lower=-10,
upper=10,
)
invProb = inverse_problem.BaseInvProblem(dmis, reg, opt)
# directives
alpha0_ratio = np.r_[1, 1, 1, 1]
alphas = directives.AlphasSmoothEstimate_ByEig(
alpha0_ratio=alpha0_ratio, n_pw_iter=10, verbose=True
)
scales = directives.ScalingMultipleDataMisfits_ByEig(
chi0_ratio=np.r_[1.0, 1.0], verbose=True, n_pw_iter=10
)
scaling_schedule = directives.JointScalingSchedule(verbose=True)
beta = directives.BetaEstimate_ByEig(beta0_ratio=1e-5, n_pw_iter=10)
beta_schedule = directives.BetaSchedule(coolingFactor=5.0, coolingRate=1)
targets = directives.MultiTargetMisfits(
TriggerSmall=False,
verbose=True,
)
# Setup Inversion
inv = inversion.BaseInversion(
invProb,
directiveList=[alphas, scales, beta, targets, beta_schedule, scaling_schedule],
)
mtik = inv.run(minit)
# Final Plot
fig, axes = plt.subplots(3, 4, figsize=(25, 15))
axes = axes.reshape(12)
left, width = 0.25, 0.5
bottom, height = 0.25, 0.5
right = left + width
top = bottom + height
axes[0].set_axis_off()
axes[0].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using true nonlinear\npetrophysical relationships"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[0].transAxes,
)
axes[1].plot(mesh.cell_centers_x, wires.m1 * mcluster_map, "b.-", ms=5, marker="v")
axes[1].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[1].set_title("Problem 1")
axes[1].legend(["Recovered Model", "True Model"], loc=1)
axes[1].set_xlabel("X")
axes[1].set_ylabel("Property 1")
axes[2].plot(mesh.cell_centers_x, wires.m2 * mcluster_map, "r.-", ms=5, marker="v")
axes[2].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[2].set_title("Problem 2")
axes[2].legend(["Recovered Model", "True Model"], loc=1)
axes[2].set_xlabel("X")
axes[2].set_ylabel("Property 2")
x, y = np.mgrid[-1:1:0.01, -4:2:0.01]
pos = np.empty(x.shape + (2,))
pos[:, :, 0] = x
pos[:, :, 1] = y
CS = axes[3].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[3].scatter(wires.m1 * mcluster_map, wires.m2 * mcluster_map, marker="v")
axes[3].set_title("Petrophysical Distribution")
CS.collections[0].set_label("")
axes[3].legend(["True Petrophysical Distribution", "Recovered model crossplot"])
axes[3].set_xlabel("Property 1")
axes[3].set_ylabel("Property 2")
axes[4].set_axis_off()
axes[4].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Using a pure\nGaussian distribution"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[4].transAxes,
)
axes[5].plot(mesh.cell_centers_x, wires.m1 * mcluster_no_map, "b.-", ms=5, marker="v")
axes[5].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[5].set_title("Problem 1")
axes[5].legend(["Recovered Model", "True Model"], loc=1)
axes[5].set_xlabel("X")
axes[5].set_ylabel("Property 1")
axes[6].plot(mesh.cell_centers_x, wires.m2 * mcluster_no_map, "r.-", ms=5, marker="v")
axes[6].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[6].set_title("Problem 2")
axes[6].legend(["Recovered Model", "True Model"], loc=1)
axes[6].set_xlabel("X")
axes[6].set_ylabel("Property 2")
CSF = axes[7].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.5,
label="True Petro. Distribution",
)
CS = axes[7].contour(
x,
y,
np.exp(clfnomapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
500,
cmap="viridis",
linestyles="--",
label="Modeled Petro. Distribution",
)
axes[7].scatter(
wires.m1 * mcluster_no_map,
wires.m2 * mcluster_no_map,
marker="v",
label="Recovered model crossplot",
)
axes[7].set_title("Petrophysical Distribution")
axes[7].legend()
axes[7].set_xlabel("Property 1")
axes[7].set_ylabel("Property 2")
# Tikonov
axes[8].set_axis_off()
axes[8].text(
0.5 * (left + right),
0.5 * (bottom + top),
("Least-Squares\n~Using a single cluster"),
horizontalalignment="center",
verticalalignment="center",
fontsize=20,
color="black",
transform=axes[8].transAxes,
)
axes[9].plot(mesh.cell_centers_x, wires.m1 * mtik, "b.-", ms=5, marker="v")
axes[9].plot(mesh.cell_centers_x, wires.m1 * m, "k--")
axes[9].set_title("Problem 1")
axes[9].legend(["Recovered Model", "True Model"], loc=1)
axes[9].set_xlabel("X")
axes[9].set_ylabel("Property 1")
axes[10].plot(mesh.cell_centers_x, wires.m2 * mtik, "r.-", ms=5, marker="v")
axes[10].plot(mesh.cell_centers_x, wires.m2 * m, "k--")
axes[10].set_title("Problem 2")
axes[10].legend(["Recovered Model", "True Model"], loc=1)
axes[10].set_xlabel("X")
axes[10].set_ylabel("Property 2")
CS = axes[11].contour(
x,
y,
np.exp(clfmapping.score_samples(pos.reshape(-1, 2)).reshape(x.shape)),
100,
alpha=0.25,
cmap="viridis",
)
axes[11].scatter(wires.m1 * mtik, wires.m2 * mtik, marker="v")
axes[11].set_title("Petro Distribution")
CS.collections[0].set_label("")
axes[11].legend(["True Petro Distribution", "Recovered model crossplot"])
axes[11].set_xlabel("Property 1")
axes[11].set_ylabel("Property 2")
plt.subplots_adjust(wspace=0.3, hspace=0.3, top=0.85)
plt.show()
Total running time of the script: (0 minutes 52.142 seconds)
Estimated memory usage: 9 MB